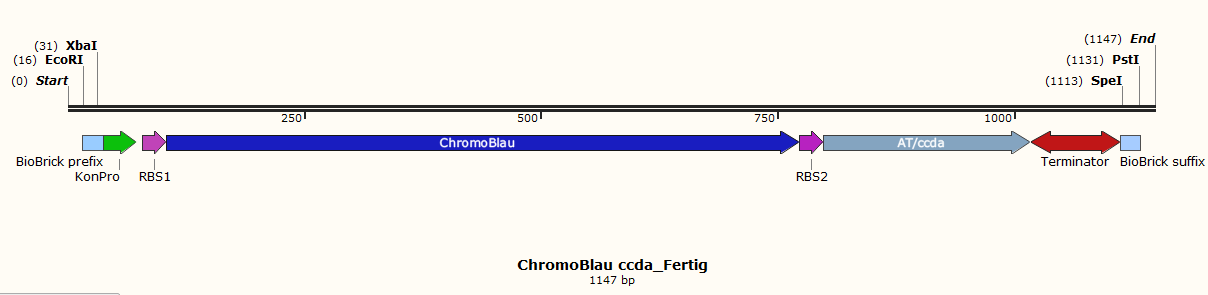
| Line 175: | Line 175: | ||
<section class="section-custom-medical"> | <section class="section-custom-medical"> | ||
| − | + | ||
| − | + | ||
| − | <div class=" | + | |
| − | + | <div class="row"> | |
| − | < | + | <div class="col-md-12"> |
| − | + | <h4 class="mb-lg">Navigation</h4> | |
| − | + | <div class="row"> | |
| + | <div class="col-md-4"> | ||
| + | <div class="tabs tabs-vertical tabs-left tabs-navigation"> | ||
| + | <ul class="nav nav-tabs col-sm-3"> | ||
| + | <li class="active"> | ||
| + | <a href="#tabsNavigation1" data-toggle="tab"><i class="fa fa-group"></i>Week 0</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation2" data-toggle="tab"><i class="fa fa-file"></i> Week 1</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation3" data-toggle="tab"><i class="fa fa-google-plus"></i> Week 2</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation4" data-toggle="tab"><i class="fa fa-adjust"></i> Week 3</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation5" data-toggle="tab"><i class="fa fa-film"></i> Week 4</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation6" data-toggle="tab"><i class="fa fa-user"></i> Week 5</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation6" data-toggle="tab"><i class="fa fa-user"></i> Week 6</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation7" data-toggle="tab"><i class="fa fa-user"></i> Week 7</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation8" data-toggle="tab"><i class="fa fa-user"></i> Week 8</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation9" data-toggle="tab"><i class="fa fa-user"></i> Week 9</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation10" data-toggle="tab"><i class="fa fa-user"></i> Week 10</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation11" data-toggle="tab"><i class="fa fa-user"></i> Week 11</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation12" data-toggle="tab"><i class="fa fa-user"></i> Week 12</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation13" data-toggle="tab"><i class="fa fa-user"></i> Week 13</a> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="#tabsNavigation14" data-toggle="tab"><i class="fa fa-user"></i> Week 14</a> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-md-8"> | ||
| + | <div class="tab-pane tab-pane-navigation active" id="tabsNavigation1"> | ||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 0</span></h2> | ||
<p> | <p> | ||
Before the actual lab work was started the whole in silico had to be planed. | Before the actual lab work was started the whole in silico had to be planed. | ||
| Line 269: | Line 323: | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation2"> | ||
| + | |||
<h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 1 (11.07 – 15.07)</span></h2> | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 1 (11.07 – 15.07)</span></h2> | ||
| Line 278: | Line 337: | ||
| − | + | </div> | |
| − | + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation3"> | |
| + | |||
| + | |||
<h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 2 (18.07 – 22.07)</span></h2> | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 2 (18.07 – 22.07)</span></h2> | ||
<p><b>Test-Transformation & pJET cloning of gBlocks</p></b> | <p><b>Test-Transformation & pJET cloning of gBlocks</p></b> | ||
| Line 289: | Line 350: | ||
| − | + | </div> | |
| − | + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation4"> | |
| + | |||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 3 (25.07 – 29.07)</span></h2> | ||
| + | <p><b>Preparing gBlocks for sequencing</b></p> | ||
| − | <p>By using the optimized blunt-end cloning protocol we finally received first positive transformation results, more specifically colonies. After doing dilution plating (5 of each strand) we also prepared ONCs out of the colonies for plasmid isolation. </p> | + | <p>By using the optimized blunt-end cloning protocol we finally received first positive transformation results, more specifically colonies. After doing dilution plating (5 of each strand) we also prepared ONCs out of the colonies for plasmid isolation. </p> |
| − | <p>Another task of the week was to prepare glucose medium, by adding glucose to our LB medium, for repression of the promotors to keep the toxin down. Otherwise they would kill themselves already in the first step of cloning. </p> | + | <p>Another task of the week was to prepare glucose medium, by adding glucose to our LB medium, for repression of the promotors to keep the toxin down. Otherwise they would kill themselves already in the first step of cloning. </p> |
| − | <p>Furthermore, at the end of the week, new diluting plating was done because the first ones did not grow in a proper way. </p> | + | <p>Furthermore, at the end of the week, new diluting plating was done because the first ones did not grow in a proper way. </p> |
| − | <img src="https://static.igem.org/mediawiki/2016/2/2d/T--NAWI-Graz--fig13.png" alt="fig13" class="img-responsive"> | + | <img src="https://static.igem.org/mediawiki/2016/2/2d/T--NAWI-Graz--fig13.png" alt="fig13" class="img-responsive"> |
| − | <h4>Fig 13: pJET Blue diluting plate (before sequencing)</h4> | + | <h4>Fig 13: pJET Blue diluting plate (before sequencing)</h4> |
| − | <img src="https://static.igem.org/mediawiki/2016/c/ce/T--NAWI-Graz--fig14.png" alt="fig14" class="img-responsive"> | + | <img src="https://static.igem.org/mediawiki/2016/c/ce/T--NAWI-Graz--fig14.png" alt="fig14" class="img-responsive"> |
| − | <h4>Fig 14: on the left side you can see the blue and the red diluting plates, | + | <h4>Fig 14: on the left side you can see the blue and the red diluting plates, |
| − | which also were working, not all of the other diluting plating were working well, just mazE.</h4> | + | which also were working, not all of the other diluting plating were working well, just mazE.</h4> |
| + | </div> | ||
| + | |||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation5"> | ||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 4 (1.08 - 5.08)</span></h2> | ||
| + | <p><b>Sequencing results and PCR</b></p> | ||
| + | <p>New ONCs of all 10 cloned gBlocks (in pJets) were done from our diluting plating. To isolate the plasmid–DNA we used the GeneJET Plasmid Miniprep Kit. To get the concentration of the extracted DNA NanoDrop was used and showed results from 22 ng/µl up to 224 ng/µl. </p> | ||
| − | + | <p>We sent 60 – 80 ng of each gBlock–pJET DNA with the proper primers for sequencing to Microsynth Austria GmbH. The following day, we analyzed the sequencing results and saw that the gBlock red, blue, lacI and mazF had the correct DNA–sequence. The other gBlocks showed no matches. Because of the fact that the other gBlocks didn`t show the expected results, we changed our plan and started to use pUC19 instead of our own designed vector and our lacI part. Unfortunately, we didn’t have enough gBlocks left for further experiments. So we decided to run a PCR, followed by a gel electrophoresis.</p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <p> | + | <p>As can be seen, the gel didn’t show clear bands at the appropriate size. </p> |
| − | < | + | <img src="https://static.igem.org/mediawiki/2016/f/fb/T--NAWI-Graz--fig15.png" alt="fig15" class="img-responsive"> |
| + | <h4>Fig 15: The sequencing results of the red part. the third clone showed the best results, so subsequent works were done with clone 3.</h4> | ||
| − | < | + | <img src="https://static.igem.org/mediawiki/2016/1/1a/T--NAWI-Graz--fig16.png" alt="fig16" class="img-responsive"> |
| + | <h4>Fig 16: This is an example how bad our sequencing results were. In the case of the vector we could work together.</h4> | ||
| − | < | + | |
| − | < | + | </div> |
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation6"> | ||
| + | |||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 5 (08.08 – 12.08)</span></h2> | ||
| + | <p><b>PCR of gBlocks for directed cloning into pUC19</b></p> | ||
| − | < | + | <p>To see the quality of our delivered gBlock–DNA, we ran a gel electrophoresis using pure gBlock-DNA. The surprising results have shown that there were some samples with very less DNA and even some with no DNA at the appropriate size. The samples that have shown a band were cut out at the correct length and resolved, followed by a PCR reaction: </p> |
| − | + | ||
| + | <p>98 °C 30 sec<br> | ||
| + | 98 °C 10 sec I<br> | ||
| + | 68 °C 20 sec I 30 x<br> | ||
| + | 72 °C 30 sec I<br> | ||
| + | 72 °C 7 min<br> | ||
| + | 4 °C ∞</p> | ||
| − | + | <p>Because of the fact that some gBlocks didn’t show results at all, we asked IDT for new samples and they sent ccdb toxin & our designed vector again. </p> | |
| − | + | ||
| − | + | ||
| − | <p> | + | <p>Moreover, a new attempt of cloning ccdB into a pJet vector was performed with the last few µl of the original sample. After transformation colonies were detected and used for a raster-screening, to double-check the size of our gBlock DNA. The correct ones were used for a colony PCR as following: </p> |
| − | <p>98 °C | + | <p>98 °C 5 min<br> |
| − | 98 °C 10 sec I<br> | + | 98 °C 10 sec I<br> |
| − | 68 °C 20 sec I | + | 68 °C 20 sec I 36 x<br> |
| − | 72 °C 30 sec I<br> | + | 72 °C 30 sec I<br> |
| − | 72 °C 7 min<br> | + | 72 °C 7 min<br> |
| − | 4 °C ∞</p> | + | 4 °C ∞</p> |
| − | + | <img src="https://static.igem.org/mediawiki/2016/d/d5/T--NAWI-Graz--fig17.png" alt="fig17" class="img-responsive"> | |
| − | + | <h4>Fig 17: The pUC19 vector we choose to work together, it has the restriction sites and the characteristics we need.</h4> | |
| − | + | </div> | |
| − | + | ||
| − | + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation7"> | |
| − | + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 6 (16.08 – 19.08)</span></h2> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <h4>Fig 17: The pUC19 vector we choose to work together, it has the restriction sites and the characteristics we need.</h4> | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<p><b>Miniprep & Sequencing of pUC19</b></p> | <p><b>Miniprep & Sequencing of pUC19</b></p> | ||
| Line 375: | Line 437: | ||
| − | + | </div> | |
| − | + | ||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation8"> | ||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 7 (22.08 – 26.08)</span></h2> | ||
<p><b>Restriction & Ligation of the parts</b></p> | <p><b>Restriction & Ligation of the parts</b></p> | ||
| Line 391: | Line 455: | ||
<p>For this reason, new restriction and ligation attempts were performed. To get a greater chance of correct colonies 3-point-ligation was done additionally. Another try was to use less PCR product. Still no result was as expected. </p> | <p>For this reason, new restriction and ligation attempts were performed. To get a greater chance of correct colonies 3-point-ligation was done additionally. Another try was to use less PCR product. Still no result was as expected. </p> | ||
| − | + | ||
| − | + | </div> | |
| + | |||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation9"> | ||
| + | |||
<h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 8 (29.08 – 02.09)</span></h2> | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 8 (29.08 – 02.09)</span></h2> | ||
<p><b>Restriction & Ligation, new ccdB & BioBricks</b></p> | <p><b>Restriction & Ligation, new ccdB & BioBricks</b></p> | ||
| Line 408: | Line 475: | ||
<p>Furthermore, our team started to particularly working on the BioBricks. It started with pJET-cloning and -transformation. </p> | <p>Furthermore, our team started to particularly working on the BioBricks. It started with pJET-cloning and -transformation. </p> | ||
| − | + | </div> | |
| − | + | ||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation10"> | ||
| + | |||
<h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 9 (05.09 – 09.09)</span></h2> | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 9 (05.09 – 09.09)</span></h2> | ||
<p><b>Restriction & Ligation, pJET & Sequencing of new ccdB</b></p> | <p><b>Restriction & Ligation, pJET & Sequencing of new ccdB</b></p> | ||
| Line 420: | Line 489: | ||
| − | + | </div> | |
| − | + | ||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation11"> | ||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 10 (12.09 – 16.09)</span></h2> | ||
<p><b>Miniprep & Protein Expression Control</b></p> | <p><b>Miniprep & Protein Expression Control</b></p> | ||
| Line 445: | Line 516: | ||
| − | + | </div> | |
| − | + | ||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation12"> | ||
| + | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 11 (19.09 – 23.09)</span></h2> | ||
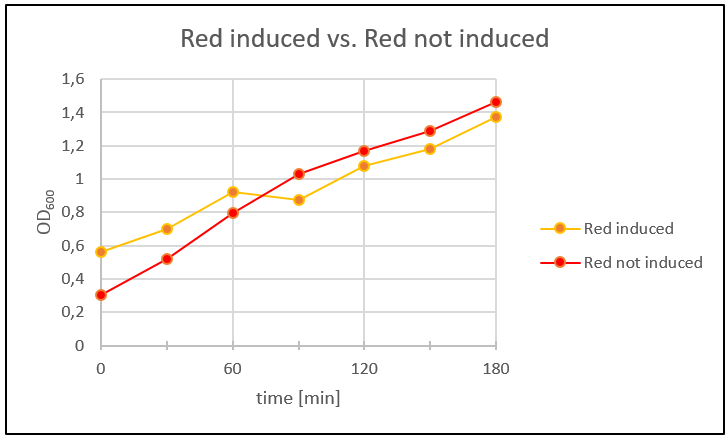
<p><b>Red_mazF & Protein Expression Control </b></p> | <p><b>Red_mazF & Protein Expression Control </b></p> | ||
| Line 481: | Line 554: | ||
<h4>Fig. 27: Eight re-streaked colonies (LB-amp plates) from a plate with Red_mazF transformants from week 10. | <h4>Fig. 27: Eight re-streaked colonies (LB-amp plates) from a plate with Red_mazF transformants from week 10. | ||
| − | + | </div> | |
| + | |||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation13"> | ||
| + | |||
<h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 12 (26.09 – 30.09)</span></h2> | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 12 (26.09 – 30.09)</span></h2> | ||
<p><b>Protein expression control in culture media</b></p> | <p><b>Protein expression control in culture media</b></p> | ||
| Line 576: | Line 652: | ||
| − | + | </div> | |
| + | |||
| + | <div class="tab-pane tab-pane-navigation" id="tabsNavigation14"> | ||
| + | |||
<h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 13 (03.10 – 07.10)</span></h2> | <h2 class="mt-xl mb-none"><span class="alternative-font font-size-md">Week 13 (03.10 – 07.10)</span></h2> | ||
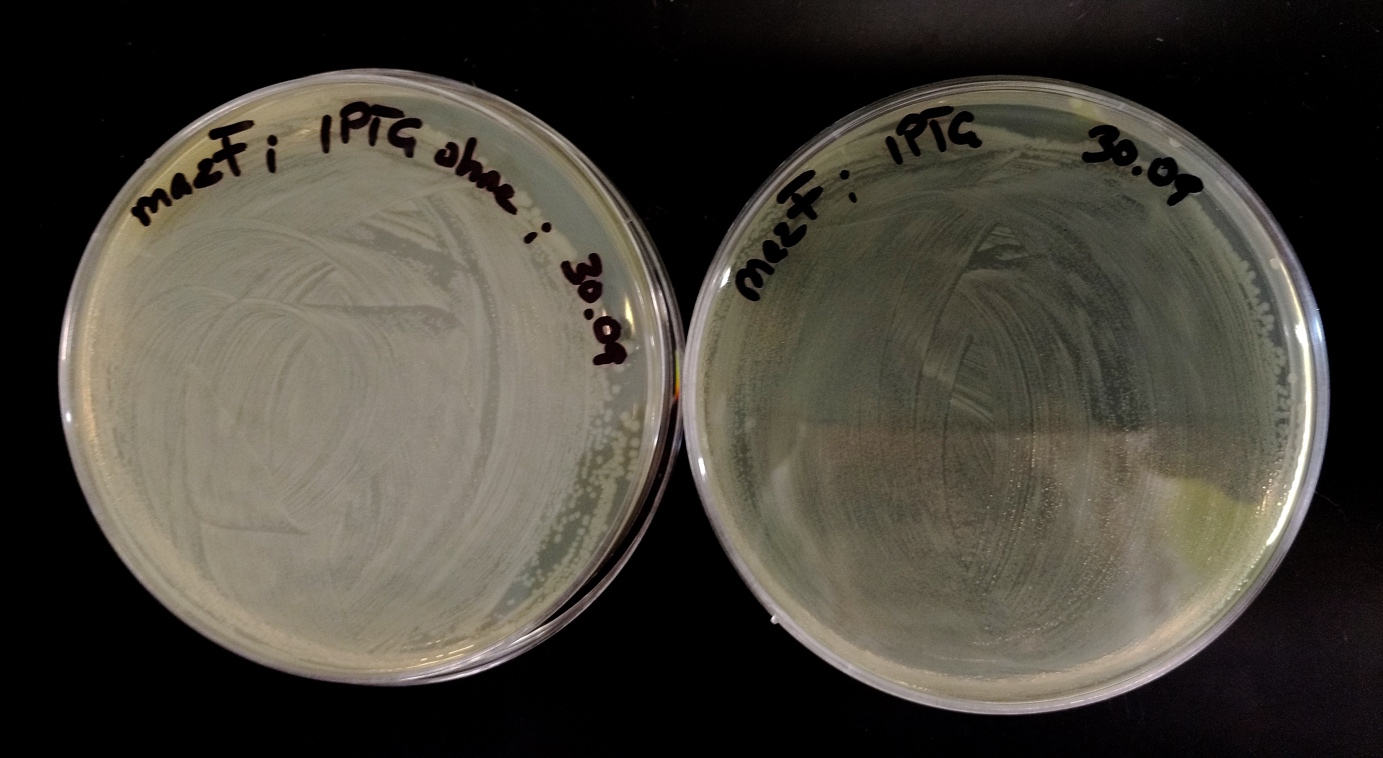
<p><b>Red_mazF vs. Blue_ccdB on LB-amp plates</b></p> | <p><b>Red_mazF vs. Blue_ccdB on LB-amp plates</b></p> | ||
| Line 605: | Line 684: | ||
<img src="https://static.igem.org/mediawiki/2016/8/84/T--NAWI-Graz--fig32.png" alt="fig32" class="img-responsive"> | <img src="https://static.igem.org/mediawiki/2016/8/84/T--NAWI-Graz--fig32.png" alt="fig32" class="img-responsive"> | ||
<h4>Fig. 32</h4> | <h4>Fig. 32</h4> | ||
| − | + | </div> | |
| + | |||
| + | |||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | ||
| + | |||
| + | |||
</section> | </section> | ||
Revision as of 09:08, 19 October 2016