Project Design
The problem we have addressed is the unprofitability of algal biofuels due to insufficient lipid content, and indirectly the greenhouse effect by promoting renewable energy. The way we have tackled this problem is by designing a construct that might enable us to control the metabolism of Chlamydomonas reinhardtii. The construct is thought to convey inducible CRISPR/Cas9 knockout of a specific metabolic target gene, directing the cells flux of carbon sources towards triacylglycerol (TAG) synthesis.
Knockout of metabolic target genes in C. reinhardtii has been successfully performed many times before. But so far, the outcome of these knockouts has not been algal strains producing enough biomass and lipids to be profitable for biodiesel production (1). For example, knocking out the sta6 gene in C. reinhardtii, encoding the starch synthesis enzyme ADP-glucose pyrophosphorylase, results in accumulation of 20 % less biomass during nutrient-replete conditions (2). However, upon nitrogen starvation, the cells turn obese. Their cytoplasm and chloroplasts fill up with TAG-bodies as a result of their increased carbon flux towards lipid synthesis (3). Even though, this knockout strain is not more productive than other strains from an overall perspective due to its reduced growth rate.
We believe the ultimate C. reinhardtii strain for biofuel production would have the growth rate of a wild type and the fatty acid accumulation of the sta6 null strain, and our hypothesis is that this could be accomplished with an inducible knockout system. By inducing the expression of Cas9 in C. reinhardtii at the biomass peak, before nitrogen starvation, more TAG-bodies would accumulate without affecting the growth rate of the algae.
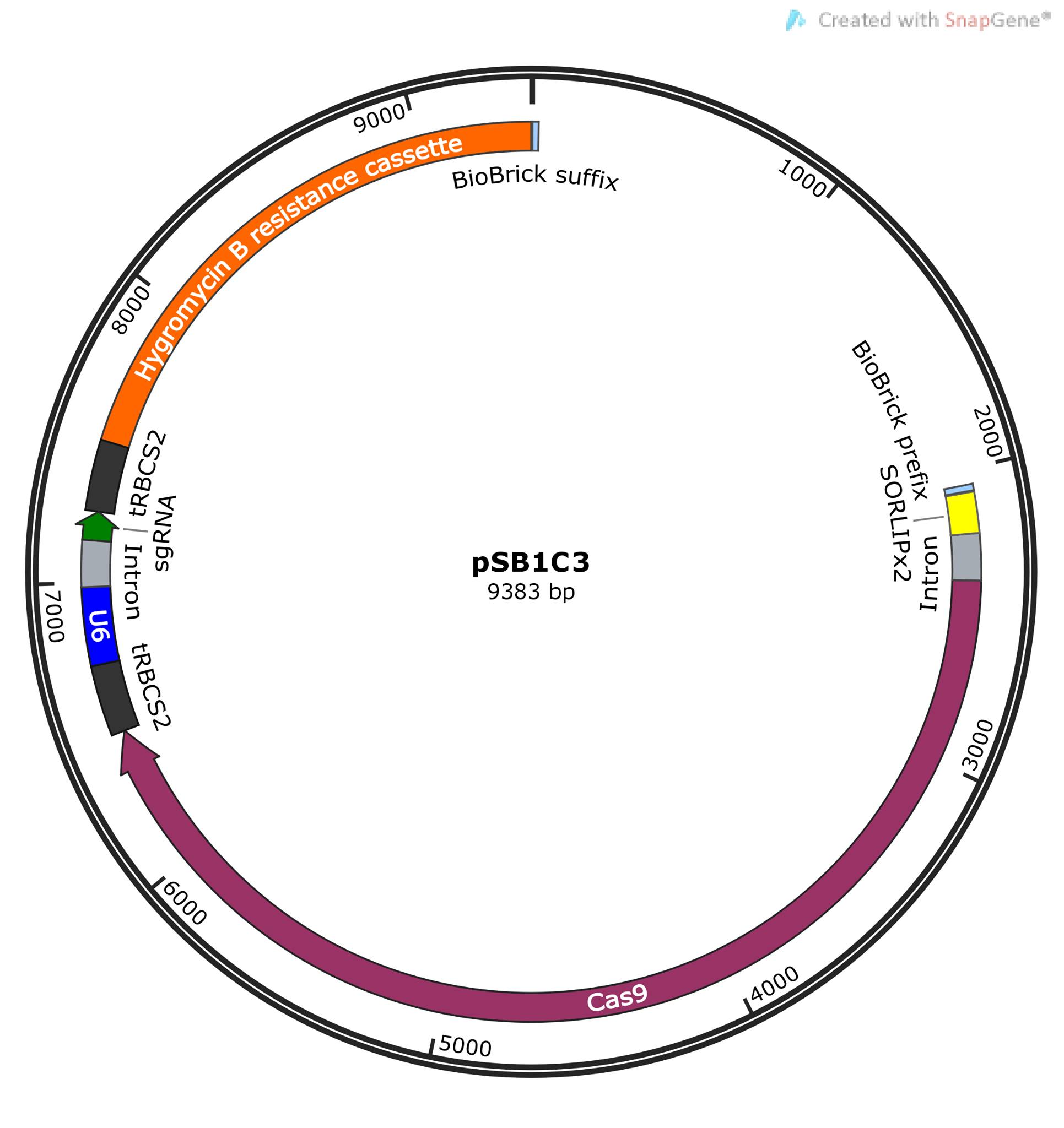
Our construct is designed to demonstrate the functionality of an inducible CRISPR/Cas9 system in C. reinhardtii. The pSB1C3 expression vector, and its chloramphenicol resistance gene, serves as base point and selection marker for cloning in E. coli. Cas9 is positively regulated by the light-inducible LIP promoter. The sgRNA, containing target sequence and scaffold sequence, is constitutively expressed under a U6-promoter. Lastly, we have a hygromycin resistance gene (aph7), serving as selection marker for successful transformation of C. reinhardtii.
The construct is constituted by six DNA fragments, assembled by Gibson assembly and designed accordingly. These are the components and their functions:
1. The high copy number expression vector pSB1C3, enabling cloning and selection in E. coli. It also provides iGEM compatibility.
2. The light inducible promotor, LIP, enables light inducible expression of Cas9 and target gene knockout. The truncated SORLIPx2 promoter sequence was retrieved from published work by Park S et al. and Baek K et al (4,5).
Turning the switch from proliferation to fatty acid accumulation on an industrial scale requires an effective stimulus. There are several variants of inducible promoters that could be used in C. reinhardtii to express a gene such as Cas9 at given stimuli, and many of these promoters include addition and removal of chemicals. Due to the potential complexity of using chemicals for this purpose, we decided to use high-intensity light as promotor stimulus. Despite that the use of a light-inducible knockout promotor in a photosynthetic organism might seem contradictory, it has its advantages due to its manageability. We also believe that a light-inducible promoter could be particularly useful for algae growing on carbon sources other than carbon dioxide, such as sugar fed or green waste algae, which do not require intensive solar energy.
3. Cas9 serves as the knockout mediator in CRISPR/Cas9 systems. The sequence used to create the fragment was retrieved from BBa_K121801 and adopted for Gibson assembly through PCR. The theoretical functionality of this part is questionable due to its lack of codon optimization for C. reinhardtii. We yet decided to use it, mainly because of the lack of other alternatives and the great effort of optimizing it ourselves, and because only a very low Cas9 expression should be required to enable target gene knockout.
4-5. sgRNA under a U6 promoter were synthesized as two fragments, based on sequences from IDT protocols (6). These fragments convey constitutive expression of sgRNA in C. reinhardtii. Since the purpose of this DNA construct was to prove the functionality of CRISPR/Cas9 in C. reinhardtii, the sgRNA does not target a metabolic gene. Instead, the sgRNA was designed to guide Cas9 to the 12-kD FK506-binding protein gene in C. reinhardtii, allowing rapamycin resistance as a selection marker for successful gene knockout.
This gene encodes the rapamycin binding protein FKBP12. A protein sensitive to rapamycin, naturally occurring in plants, including the algae of interest, C. reinhardtii (7). Rapamycin is an antibiotic which inhibits cell growth. It acts by binding to FKBP12, and the rapamycin-FKBP12 complex then inhibits the target of rapamycin (TOR). The normally occurring, active form of TOR frames the cell growth of the algae, and its inhibition induces cell death. Consequently, knock out of this gene mediates rapamycin resistance – allowing colonies to survive and grow on rapamycin plates (7).
6. The function of the last fragment is to enable selection of successfully transformed colonies of C. reinhardtii. It includes of a codon optimized hygromycin resistance gene, Aph7, which can be used as a selection marker (8). The fragment we designed is equivalent to BBa_K1857001. Unfortunately, the BioBrick was unavailable but the cassette could be retrieved through PCR from the Popt_mVenus_Hyg expression plasmid, provided by Chlamydomonas Resource Center. This could successfully be achieved but one problem in the design was that the region of interest in Popt_mVenus_Hyg, was not unique. Our primers also isolated the mVenus part of the plasmid, a yellow fluorescent protein (YFP) in almost exactly same size as the hygromycin-casette fragment. The only way to separate these fragments was by sequencing. Theoretically, the YFP reporter gene could also serve as a selection marker, therefore we continued with the primer design.
Despite multiple attempts, the assembly has not been fully successful, yet. An insert that appeared to be correct was transformed into C. reinhardtii but selection failed, and the insert later proved to be wrong (see results).
References
1. Goncalves EC, Wilkie AC, Kirst M, Rathinasabapathi B. Metabolic regulation of triacylglycerol accumulation in the green algae: Identification of potential targets for engineering to improve oil yield. Plant Biotechnology Journal. 2016;
2. Krishnan A, Kumaraswamy GK, Vinyard DJ, Gu H, Ananyev G, Posewitz MC, et al. Metabolic and photosynthetic consequences of blocking starch biosynthesis in the green alga Chlamydomonas reinhardtii sta6 mutant. Plant J. 2015;81(6):947–60.
3. Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, et al. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell [Internet]. 2014 May [cited 2016 Sep 18];13(5):591–613. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24585881
4. Park S, Lee Y, Lee J-H, Jin E. Expression of the high light-inducible Dunaliella LIP promoter in Chlamydomonas reinhardtii. Planta [Internet]. 2013 Dec [cited 2016 Sep 4];238(6):1147–56. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24043576
5. Baek K, Lee Y, Nam O, Park S, Sim SJ, Jin E. Introducing Dunaliella LIP promoter containing light-inducible motifs improves transgenic expression in Chlamydomonas reinhardtii. Biotechnol J. 2016;11(3):384–92.
6. IDT. CRISPR/Cas9 editing: transfection of gBlocks® Gene Fragments that encode sgRNAs. 2015 [cited 2016 Oct 7];4. Available from: http://eu.idtdna.com/pages/docs/default-source/user-guides-and-protocols/gblocks-gene-fragment-transfection.pdf?sfvrsn=6
7. Crespo JL, Díaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol [Internet]. 2005 Dec [cited 2016 Sep 4];139(4):1736–49. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16299168
8. Lauersen KJ, Kruse O, Mussgnug JH. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl Microbiol Biotechnol [Internet]. 2015 Apr [cited 2016 Sep 4];99(8):3491–503. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25586579