Abstract
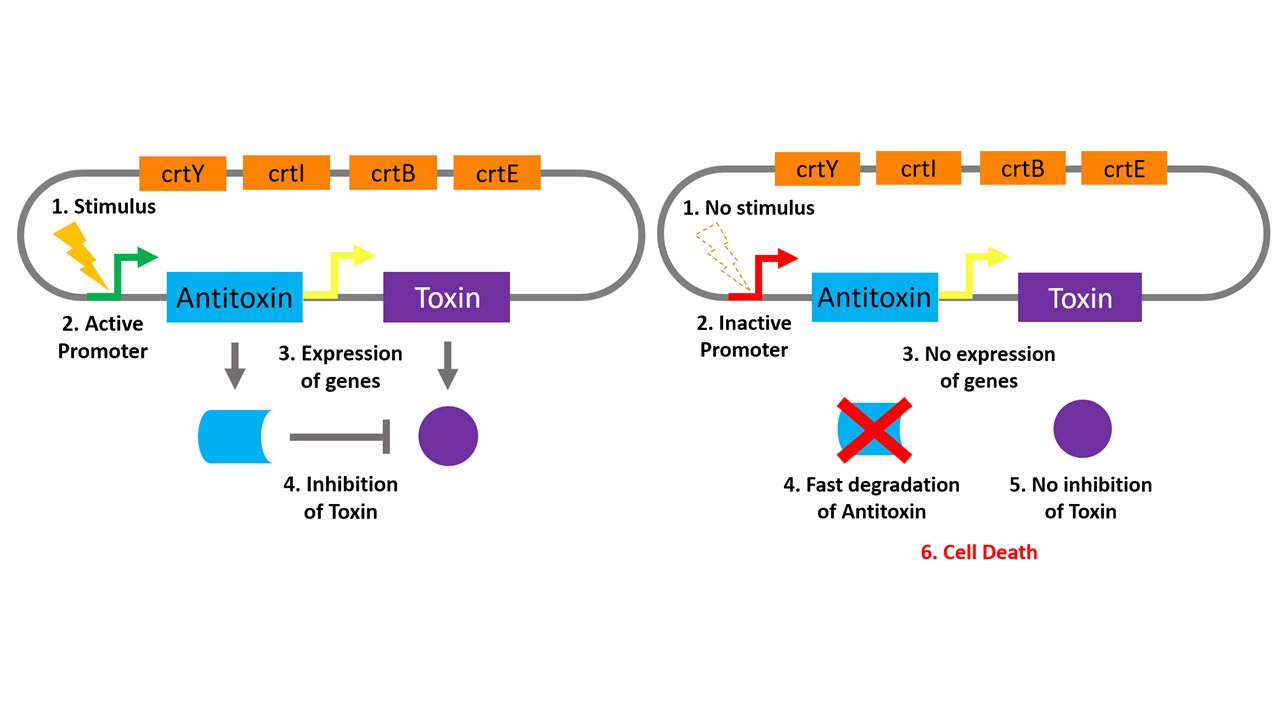
To ensure plasmid retention and prevent the release of our modified bacteria into the environment, we aimed to create a self-inactivation mechanism (SIM). This would consist of a modified toxin-antitoxin (TA) addiction system, used in natural systems by low-copy number plasmids to ensure post-segregational killing of daughter cells lacking the plasmid [1]. TA systems are based on the expression of two genes: a toxin and an antitoxin. TA systems are tightly linked to avoid mutation and so exist on the same plasmid, under the same promoter, often with overlapping sequences. The toxin has a longer half-life than the antitoxin, therefore, if a daughter cell does not inherit the plasmid and express the antitoxin, then the lingering antitoxin protein will be degraded before the toxin leaving enough of the toxin to kill the cell. Our plan was to modify this system to create a plasmid in which the toxin is constitutively expressed and the antitoxin is only expressed when in the milk/yogurt environment. In this way the bacteria are unable to escape the target environment without losing expression of the antitoxin and becoming inactivated. This is the basis of our SIM, and can be seen in the diagram below in which the stimulus is the milk/yogurt environment.
The toxin-antitoxin system we chose to use is the epsilon-zeta system from plasmid pSM19035 from Streptococcus pyogenes, in which epsilon is the antitoxin that binds to and inactivates zeta. We assembled epsilon in pSB1C3 under the control of an Anderson promoter (J23106). We were unable to transform E.coli with zeta without a promoter into pSB1C3 or into pSB3K3 (a low-copy number plasmid). Lacking a promoter and in a low copy number plasmid, we hoped to have as little expression of zeta as possible while not in a plasmid construct with epsilon. We planned to use qRT-PCR to measure the expression of epsilon but sadly ran out of time. From a human practices perspective, we talked to John Walls, an expert in public perception of synthetic biology, and discussed whether people were more likely to consume yogurt containing GMOs with the knowledge that the bacteria would not survive in their body and also that the use of antibiotics to continually select for transformed bacteria in the yogurt would not be needed. It is not ideal to use antibiotic selection as this encourages the culturing of superbugs – harmful bacteria resistant to antibiotics.
Background
Genetically modified organisms are generally contentious, especially those that are intended to be consumed [2]. To reduce this barrier to use, we engineered a self-inactivation mechanism (SIM) that inactivates our GM bacteria on an environmental basis. Since we intend to use our bacteria to generate vitamin-enriched yogurt, we wanted the SIM device to inactivate the organisms when they are out with the milk/yogurt environment. This could make our system more acceptable for the public in the sense that any GM organisms would not be able to survive outside of the product. We hope that this may reduce anxiety surrounding the use and consumption of genetically modified organisms.
Not only will the SIM device be an asset to our product from a human perspective, it will also help with retention of the vitamin producing properties encoded on the plasmid used in this genetic modification. Our SIM has a dual effect because we utilised a toxin-antitoxin (TA) system, used in nature as a plasmid retention system for low-copy number plasmids [1]. The main alternative to using a TA system for plasmid selection is antibiotic resistance. We wanted to avoid this selection method for three main reasons. 1. due to the crisis of antibiotic resistance 2. we did not want our consumers to be faced with reduced bacterial diversity in their gut as a consequence of consuming our yogurt 3. the product should be sustainable for people in resource limited settings, with limited access to antibiotics [3]
Antibiotic resistance is where repeated use of antibiotics provides a selective pressure for bacteria to develop a mechanism to survive the antibiotic, either by breaking it down, pumping it out of the cell or modifying or removing its target [3]. The issue currently is that antibiotics are being overprescribed and this has resulted in increased selection and a rise in antibiotic resistant bacteria. For disease causing (pathogenic) bacteria, antibiotic resistance causes problems as treatments rely on antibiotics and in some cases there are few antibiotics available for that particular infection. In using an antibiotic to select for the modified bacteria in our yogurt, we would be subjecting the guts of consumers to this selective pressure as they ingest the antibiotic, possibly resulting in the rise of pathogenic, antibiotic resistant bacteria from the consumers’ natural flora (the bacteria that naturally live in the gut). This is a problem as the antibiotic resistant bacteria in the natural flora would be able to outcompete non-resistant bacteria which may be of benefit to the host, and also if pathogenic they can cause illness in the host. Using antibiotic selection massively reduces the diversity of bacteria in a given environment in that only those carrying resistance genes will be able to survive. When ingested, the yogurt would apply this kind of selective pressure on the gut natural flora and so would reduce the bacterial diversity [4] within the consumer’s gut. For these reasons and the lack of distribution of antibiotics in poorer countries, we chose our modified TA system to be our method of plasmid retention as it is a system that keeps the plasmid selected for within cells without the need for antibiotics.
TA systems are based on the expression of two genes; one encoding a toxin and the other encoding an antitoxin. The common feature of TA systems is that the toxin has a longer half-life than the antitoxin [1]. In this way, if the plasmid is lost, the antitoxin with a shorter half-life will be degraded and the toxin will be free to either kill the cell or prevent growth. Generally, there are broadly two forms of TA-system; RNA based and protein based [5]. We opted to use a protein based system since RNA based systems are highly integrated and therefore are difficult to separate. This is due to overlapping coding sequences, for example with the hok-sok system, and where our project involves placing the different components of the TA system under separate promoters (to ensure survival within a yogurt/milk environment only), this would not be possible with a system where the sequences are tightly connected.
The specific toxin-antitoxin system that we use comes from Streptococcus pyogenes - a close relative of Streptococcus thermophiles, which is at the basis of our yogurt. It is contained on the plasmid pSM19035 and is comprised of three main genes referred to as epsilon, omega and zeta. Epsilon encodes the antitoxin, zeta encodes the toxin and omega is thought to encode an auto regulatory protein that aids the system by regulating transcription. As our system involves placing epsilon and zeta under synthetic promoters, omega should not be necessary. Epsilon inhibits zeta through the binding of its N-terminal domain with zeta’s N-terminal domain, which is responsible for its toxicity [6]. When epsilon is bound to zeta, zeta is inactive and does not kill the cell.
Lab Work
We identified these three genes and set out to experiment with them in E. coli. We used the sequences found in the NCBI gene database for the natural coding sequence for all three genes [7] without a ribosome binding site (RBS) as well as all three with the native RBS and codon bias towards GC content. The genes were ordered without an RBS at first, however due to the low GC content, the manufacturer was having difficulties synthesising them. We thus reordered them but with a more favourable GC content and with their natural strength RBS. Using a RBS strength calculator [8], we found that the strength of RBS is around 72.5 times stronger for Epsilon than for Zeta as the antitoxin must be made in excess of the toxin. When we found this strength difference, we decided that the sequences without an RBS would be less useful than those ordered with the natural strength RBS with the great difference in strength.
To test both parts of the toxin-antitoxin system, we aimed to insert epsilon and zeta individually into pSB1C3 under the pBAD promoter (I0500) in separate cells. Using this, we planned to add arabinose to show that epsilon does not kill the cells and see its impact on the growth rate through OD. When expression of zeta is induced, we would have expected to see the cells die. In practice, we were unable to transform cells with zeta in the plasmid pSB1C3 without a promoter, as either there is no growth on plates which should have transformants or colonies that have been mini-prepped from these transformations have lacked the plasmid. The former may have been due to run-through transcription resulting in expression of zeta which is then free to kill the cells after transformation, meaning the plates will show lack of growth of colonies. We inserted zeta into a low-copy number plasmid pSB3K3 with pBAD (as a single ligation step), and used electroporation (to increase transformation efficiency). The cells were left to grow overnight with 0.2% D-glucose added to the media to ensure the pBAD promoter was as inactive as possible, as pBAD is induced by L-arabinose but inhibited by glulcose [9]. However, this failed to work as well. We successfully assembled pSB1C3 with epsilon behind an Anderson promoter (J23106). We did not have time to test this part http://parts.igem.org/Part:BBa_K2151102 BBa_K2151102.
Our strategy was to go through 3 steps:
- a test system, where epsilon expression would be under the control of a constitutive promoter and zeta under an inducible promoter. This should have shown that epsilon is not toxic to the cells and that inducing zeta does not kill the cells when epsilon is present.
- a final lab system, where epsilon expression would be under an inducible promoter and zeta under a constitutive promoter. This should have shown that when epsilon expression is induced, the cells survive, but when the pBAD is no longer induced, zeta would kill the cells.
- a real-world system, where epsilon expression would be under a promoter identified through transcriptomics to be active only in the milk/yogurt environment. Under this system, the bacteria would be able to survive in the target environment (milk/yogurt) but would be unable to be released into the general environment or survive inside people who have ingested the yogurt as well as facilitating plasmid retention. (the transcriptomics results are presented here)
Discussion
Zeta (the toxin) is a bactericidal protein that affects peptidoglycan synthesis so can also have affects in eukaryotes like S. cerevisiae [10], however it is not harmful to human cells as we do not use peptidoglycan. This makes this particular TA system fit for our purpose as the bacteria with the toxin and antitoxin could be ingested without harm.
Given more time to work on this project, we would like to carry out all the testing we aimed to complete as mentioned earlier. To do this we would have to successfully transform cells with; zeta in pSB3K3 with the pBAD promoter, epsilon in pSB1C3 with a pBAD promoter and epsilon behind a pBAD promoter and zeta behind a constitutive promoter in pSB1C3. Not only could our SIM be used in products to be ingested or more generally that could be in contact with the human body, it could potentially be used in the lab as an experimental tool for triggered cell death under the regulation of chosen promoters and as a mechanism to prevent harmful bacteria from growing outside of the lab. This latter point was a key component of our work on human practices.
References
- ↑ 1.0 1.1 1.2 Zielenkiewicz, U. and P. Ceglowski. "The Toxin-Antitoxin System Of The Streptococcal Plasmid Psm19035". Journal of Bacteriology 187.17 (2005): 6094-6105.
- ↑ Marris, Claire. "Public Views On Gmos: Deconstructing The Myths". EMBO reports 2.7 (2001): 545-548.
- ↑ 3.0 3.1 Carlet, Jean and Didier Pittet. "Access To Antibiotics: A Safety And Equity Challenge For The Next Decade". Antimicrobial Resistance & Infection Control 2.1 (2013): 1.
- ↑ Guarner, Francisco and Juan-R Malagelada. "Gut Flora In Health And Disease". The Lancet 361.9356 (2003): 512-519.
- ↑ Brantl, Sabine. "Bacterial Type I Toxin-Antitoxin Systems". RNA Biology 9.12 (2012): 1488-1490.
- ↑ Zielenkiewicz, U. et al. "In Vivo Interactions Between Toxin-Antitoxin Proteins Epsilon And Zeta Of Streptococcal Plasmid Psm19035 In Saccharomyces Cerevisiae". Journal of Bacteriology 191.11 (2009): 3677-3684.
- ↑ Unterholzner, Simon J, Brigitte Poppenberger, and Wilfried Rozhon. "Toxin–Antitoxin Systems". Mobile Genetic Elements 3.5 (2013): e26219.
- ↑ "Salis Lab: The Ribosome Binding Site Calculator". Salislab.net. N.p., 2016. Web. 14 Oct. 2016.
- ↑ Guzman, Luz-Maria et al. "Tight Regulation, Modulation, And High-Level Expression By Vectors Containing The Arabinose PBAD Promoter". Journal of Bacteriology 177.14 (1995): 4121–4130.
- ↑ Yeo, Chew et al. "Heterologous Expression Of Toxins From Bacterial Toxin-Antitoxin Systems In Eukaryotic Cells: Strategies And Applications". Toxins 8.2 (2016): 49.