Existing parts
Basic
| Name | Type | Designer | Length |
|---|---|---|---|
| BBa_R0040 | pTet promoter | June Rhee, Connie Tao, Ty Thomson, Louis Waldman Group: Antiquity (2003-01-31) | 54 bp |
Composite
| Name | Type | Designer | Length |
|---|---|---|---|
| BBa_K1352000 | Ag43 under pBAD/araC promoter | June Rhee, Connie Tao, Ty Thomson, Louis Waldman Group: Antiquity (2003-01-31) | 54 bp |
| BBa_I746908 | sfGFP under pBAD/araC promoter | Ana-Maria Cujba, Martyna Sroka Group: iGEM14_Aberdeen_Scotland (2014-10-01) | 4493 bp |
| BBa_K1072000 | Lux based quorum sensing system | Yichong wang, Rui chen, Xihao liao and Junjie zhang Group: iGEM13_SCUT (2013-08-31) | 2910 bp |
| BBa_K525998 | This part consists of a T7 bacteriophage promoter followed by a very strong RBS – BBa_B0034 | Anna Drong Group: iGEM11_Bielefeld-Germany (2011-09-13) | 32 bp |
Description
BBa_R0040
This part was used as a negative control for fluorescence measurements, since it is non-coding except for chloramphenicol resistance.
BBa_K1352000Ag43 under pBAD/araC promoter. This part codes with the Ag43 pro-protein under the control of the pBAD/araC regulation system.
Briefly, Antigen 43 (Ag43) is a bacterial cell adhesion outer membrane protein, belonging to the auto-transporter family. It consists of two subunits – β, a trans-membrane β-barrel protein and α, a secretory protein that non-covalently binds to the β subunit and is responsible for the aggregation by forming α-α dimers. These two subunits come from the same peptide chain that is cleaved by an aspartate protease domain in the same pro-protein. Phenotypically, Ag43 expression leads to inter-cell aggregation.
We used BBa_K1352000 as PCR templates to make tagged variants of Ag43 (BBa_K2168000 and BBa_K2168001). For a excellent description of Ag43, we recommend visitingThis part codes for sfGFP under regulation of pBAD/araC system.
We used this part to characterize the pBAD/araC regulation system, more specifically promoter activity repression by glucose, since this had paradoxically not been checked in the past 12 years. For more details about experiments and results, please click
Lux based quorum sensing system.
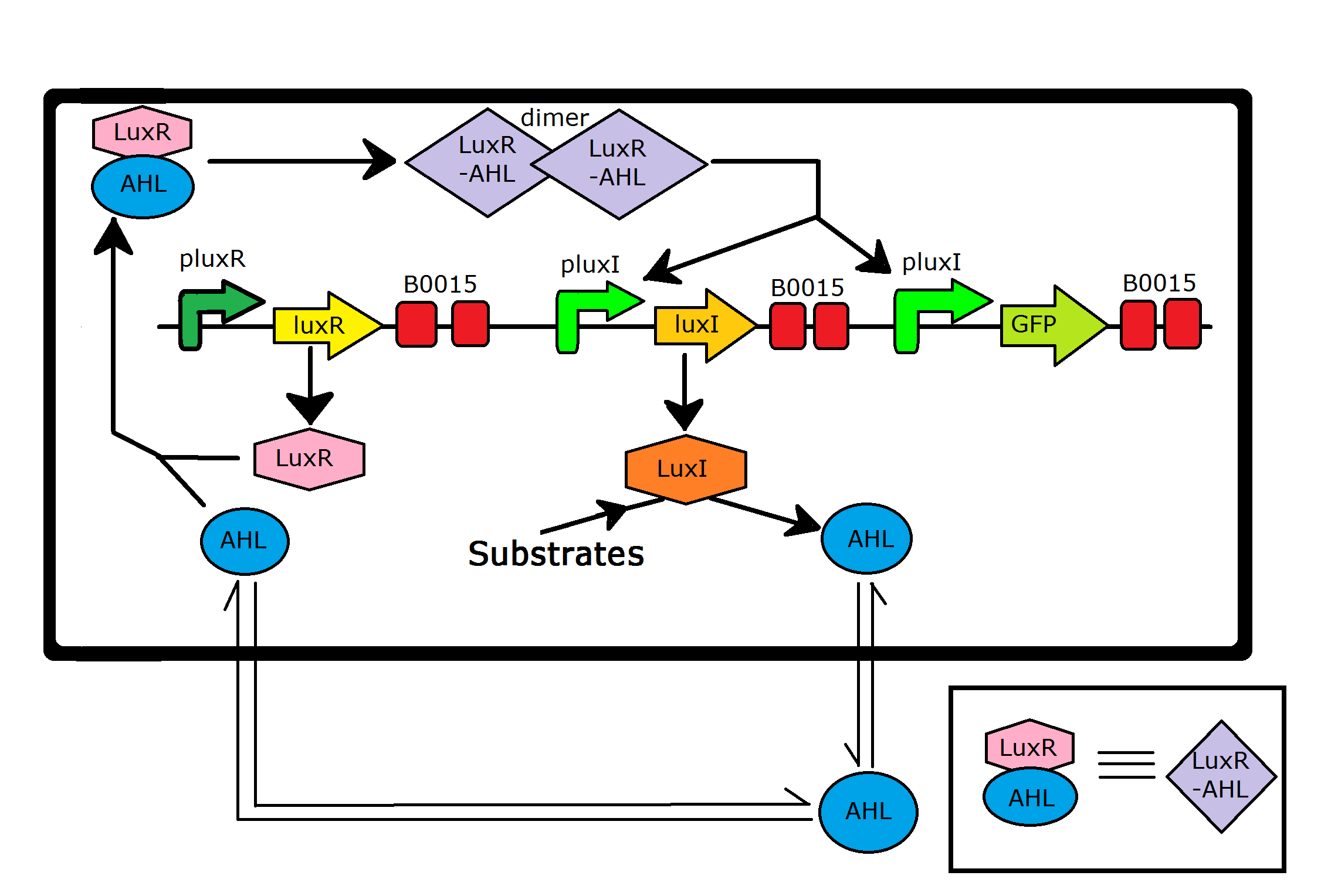
Quorum sensing systems are biological regulation circuits that allow gene regulation dependent on local cell-densities. A well-studied example is that in Aliivibrio fischeri lux quorum sensing system.
The system works as follows. luxI gene under the pLuxI promoter encodes for the LuxI AHL-synthetase enzyme that synthesizes AHL, non-polar signalling molecules that can diffuse out into the extracellular environment. Another protein encoded by this part, LuxR from the luxR gene under the pluxR promoter, binds to AHL and the LuxR-AHL complex homodimerizes further to give a (LuxR-AHL) dimer. This dimer acts as a transcriptional activator for the pluxI promoter, leading to a positive feedback loop for luxI-luxR expression and AHL synthesis.
Further, the dimerization of the LuxR-AHL complex leads to a positive Hill coefficient for the process leading to a switch-like behaviour with respect to the AHL concentration. This lead to a switch like behaviour with respect to cell-density since the AHL production rate is dependent on the local cell density.
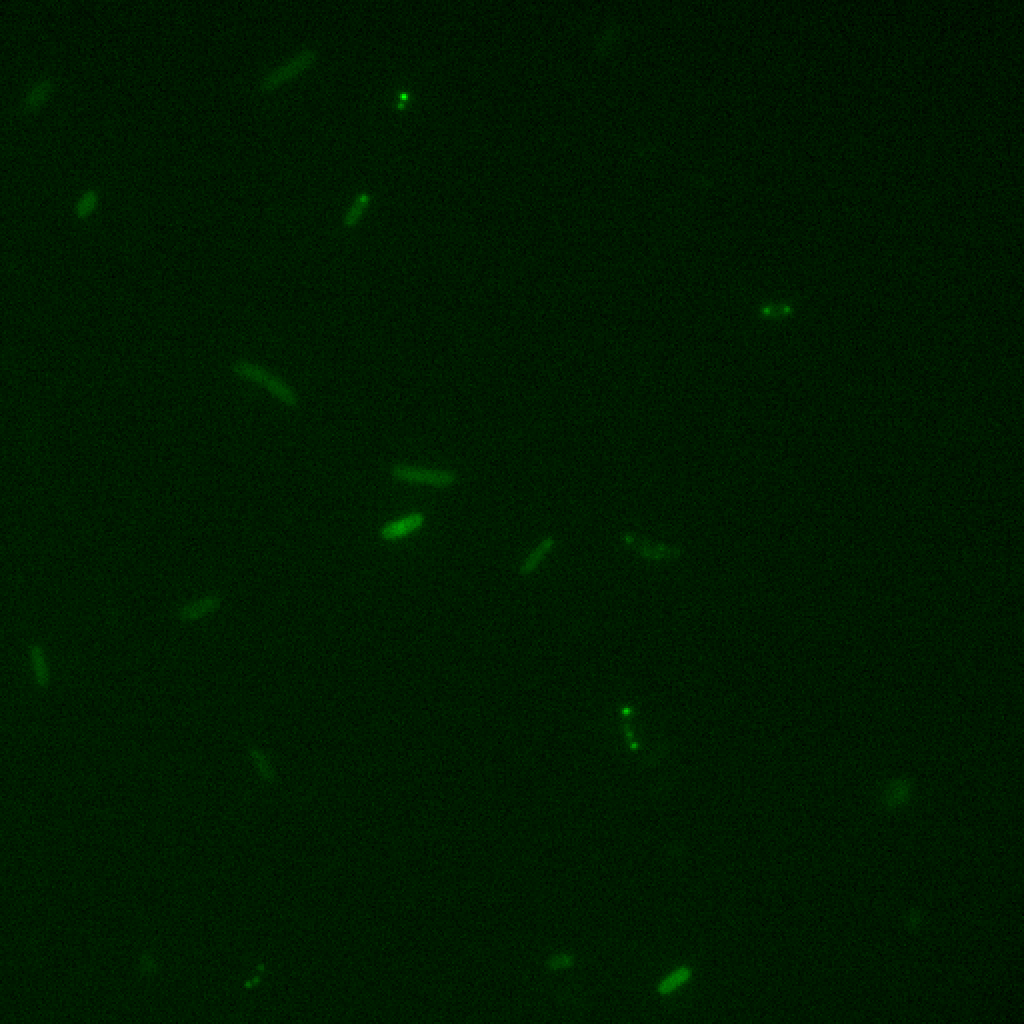
Hence, any gene under the pluxI promoter would show a switching behaviour dependent on cell-density. We decided to check the same for BBa_K1072000, one for the few parts that has a complete quorum sensing system with a fluorescent reporter. So far, the characterization of this part has a microscopy image of green fluorescent cells; however, quorum sensing can established only by a growth curve. We have shown quorum sensing by this part experimentally. For experimental details, please click
This part consists of a T7 bacteriophage promoter followed by a very strong RBS – BBa_B0034. This part was chosen by us for collaborating with the
New parts
| Name | Type | Designer | Length |
|---|---|---|---|
| BBa_K2168000 | Arunavo Chakraborty Group: iGEM16_IISc_Bangalore (2016-10-13) | 4511 bp | |
| BBa_K2168001 | Arunavo Chakraborty Group: iGEM16_IISc_Bangalore (2016-10-13) | 5258 bp |
This is a modification of BBa_K1352000, where Ag43 has been tagged on the N-terminal with a 6xHis tag for purification and quantification. This part was prepared by Gibson assembly, the details of which can be found
This is a modification of BBa_K1352000, where Ag43 has been tagged at the N-terminal with 6xHis tag and at the C-terminal with a sfGFP tag. This part was prepared by Gibson assembly, the details of which can be found
here.
In addition to the location of the 6xHis tag, the linker between the Ag43 and the sfGFP coding sequences needs to be chosen with some thought. The linker that we choose as a (GGGGS)2 amino acid sequence, which would be a flexible linker with intermediate hydrophobicity.
However, there is a reasonable possibility of this part being non-functional since the C-terminal amino acid for Type V auto-transporter family (of which Ag43 is a member) seems to be critical for integration of the protein on the outer-membrane. Since the C-terminal of Ag43-sfGFP would no longer be free, issues with aggregation due to BBa_K2168001 were expected.
Experimentally, we found the over-expressed Ag43-sfGFP chimera to be present in cellular interior based on epifluorescence microscopy images of induced cells.

Parts collection
Ag43 measurement systems
| Name | Type | Designer | Length | |
|---|---|---|---|---|
| BBa_K2168000 | Arunavo Chakraborty Group: iGEM16_IISc_Bangalore (2016-10-13) | 4511 bp | ||
| BBa_K2168001 | sfGFP under pBAD/ara promoter | Arunavo Chakraborty Group: iGEM16_IISc_Bangalore (2016-10-13) | 5258 bp |
Planned parts
Native lux system:
Given our interest in protein-over-expression, we designed a quorum sensing system to allow for greater protein-over-expression without leaky expression. For this, we hypothesized that have bicistronic expression system for luxI and GFP, since it decreases the competition for LuxR-AHL dimer by the pluxI. So, we ordered the native lux system codon optimized for E coli from IDT, however, we received empty tubes from them.
Overview
With the advent of rDNA technology in the late 1970s, medicine, agriculture and several other areas underwent a quantum leap and from that point, progress only hastened, from one only one recombinant pharmaceutical approved for human use (insulin) in 1982 to one hundred and fifty-one FDA approved protein based recombinant pharmaceuticals by 2009[1].
Despite being in high demand (due to the fact that most recombinant products produced on an industrial scale are therapies for chronic diseases like cancer and diabetes), recombinant products are expensive due to several factors like long and expensive development time, high failure rate (~80%) of the products developed, manufacturing costs requiring expensive technologies and processes (bioreactors, column chromatography, sterile conditions, etc) and the involvement of skilled labor on both the manufacturing and the healthcare provider’s side[2]. Treatments with these pharmaceuticals can cost from around 10,000 to 100,000 € per year for a single patient[2]. As scientists and engineers, it seems obvious that our contribution can be most easily and effectively be made at the level of manufacturing costs; to try to bring down the cost of these life-saving products.
Read more








