| Line 98: | Line 98: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div class="p_font_size"><small>Figure 10 The biosynthesis pathway of astaxanthin formation in transgenic rice endosperm. The dotted arrows indicate pathway limitations in rice endosperm. The solid arrows indicate the existence of carotenogenic reactions. The red arrows indicate the reactions catalaysed by four exogenous transgenes Psy, CrtI, BHY and BKT.</small></div> | + | <div class="p_font_size" style="margin-bottom:20px"><small>Figure 10 The biosynthesis pathway of astaxanthin formation in transgenic rice endosperm. The dotted arrows indicate pathway limitations in rice endosperm. The solid arrows indicate the existence of carotenogenic reactions. The red arrows indicate the reactions catalaysed by four exogenous transgenes Psy, CrtI, BHY and BKT.</small></div> |
| − | <div class="row | + | <div class="row"> |
<div class="col s12" align="center"> | <div class="col s12" align="center"> | ||
<img alt="image" class="img-responsive col s11" src="https://static.igem.org/mediawiki/2016/4/42/T--SCAU-China--model_p2.png"> | <img alt="image" class="img-responsive col s11" src="https://static.igem.org/mediawiki/2016/4/42/T--SCAU-China--model_p2.png"> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div class="p_font_size"><small>Figure 11 Correlation analyses between expression levels of four genes involved in astaxanthin biosynthesis and astaxanthin content.</small></div> | + | <div class="p_font_size" style="margin-bottom:20px"><small>Figure 11 Correlation analyses between expression levels of four genes involved in astaxanthin biosynthesis and astaxanthin content.</small></div> |
<br> | <br> | ||
| Line 116: | Line 116: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div class="p_font_size">The change in concentration of protein Y is due to the difference between its production and degradation/dilution, as described by a dynamic equation:<div> | + | <div class="p_font_size">The change in concentration of protein Y is due to the difference between its production and degradation/dilution, as described by a dynamic equation:</div> |
<div class="row"> | <div class="row"> | ||
<div class="col s12" align="center"> | <div class="col s12" align="center"> | ||
| Line 191: | Line 191: | ||
<div class="p_font_size">In the first step of our modeling, we support that all genes have the same expression way in the cell. We support that all the input signals appear at time t=0. Because we don’t have enough time to measure transcription and translation rates of the genes, we assume that all the proteins have the same production rate and this rate are supported to be 200 nM/h. To measure the degradation rate of protein, αdeg, T1/2 is used in the following calculation, in which period of time the protein decrease by 50%.T1/2 of each protein is determined by their Instability index (II). Degradation rates of protein can be calculated by the aquation, T1/2 =log(2)/α.Yst means steady-state concentration of the proteins.</div> | <div class="p_font_size">In the first step of our modeling, we support that all genes have the same expression way in the cell. We support that all the input signals appear at time t=0. Because we don’t have enough time to measure transcription and translation rates of the genes, we assume that all the proteins have the same production rate and this rate are supported to be 200 nM/h. To measure the degradation rate of protein, αdeg, T1/2 is used in the following calculation, in which period of time the protein decrease by 50%.T1/2 of each protein is determined by their Instability index (II). Degradation rates of protein can be calculated by the aquation, T1/2 =log(2)/α.Yst means steady-state concentration of the proteins.</div> | ||
| − | + | <table class="table"> | |
| − | + | <thead> | |
| − | + | <th>Protein names</th> | |
| − | + | <th>Gene names</th> | |
| − | + | <th>Organism</th> | |
| − | + | <th>Instability index (II) </th> | |
| − | + | <th>β</th> | |
| − | + | <th>αdeg</th> | |
| − | + | <th>Yst</th> | |
| − | + | </thead> | |
| − | + | <tbody> | |
| − | + | <tr> | |
| − | + | <td>Actin</td> | |
| − | + | <td>OsAct1</td> | |
| − | + | <td>Oryza sativa subsp. japonica</td> | |
| − | + | <td>34.96</td> | |
| − | + | <td>200 nM/h</td> | |
| − | + | <td>5.26 </td> | |
| − | + | <td>1</td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td>phytoene synthase</td> | |
| − | + | <td>PSY</td> | |
| − | + | <td>Zea mays L.</td> | |
| − | + | <td>59.82</td> | |
| − | + | <td>200 nM/h</td> | |
| − | + | <td>9.09 </td> | |
| − | + | <td>1.73</td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td>phytoene desaturase</td> | |
| − | + | <td>CrtI/PDS</td> | |
| − | + | <td>Erwinia uredovora</td> | |
| − | + | <td>31.38</td> | |
| − | + | <td>200 nM/h</td> | |
| − | + | <td>4.69 </td> | |
| − | + | <td>0.89</td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td>β-carotene hydroxylase</td> | |
| − | + | <td>BHY</td> | |
| − | + | <td>Haematococcus pluvialis</td> | |
| − | + | <td>52.32</td> | |
| − | + | <td>200 nM/h</td> | |
| − | + | <td>7.89 </td> | |
| − | + | <td>1.50</td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td>β-carotene ketolase</td> | |
| − | <div class="p_font_size"> | + | <td>BKT</td> |
| − | + | <td>Chlamydomonas reinhardtii</td> | |
| + | <td>40.90</td> | ||
| + | <td>200 nM/h</td> | ||
| + | <td>6.12 </td> | ||
| + | <td>1.16</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>phytoene desaturase</td> | ||
| + | <td>PDS</td> | ||
| + | <td>Oryza sativa cv.</td> | ||
| + | <td>49.69</td> | ||
| + | <td>200 nM/h</td> | ||
| + | <td>7.5 </td> | ||
| + | <td>1.43</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>ζ-carotene desaturase</td> | ||
| + | <td>ZDS</td> | ||
| + | <td>Oryza sativa cv.</td> | ||
| + | <td>49.12</td> | ||
| + | <td>200 nM/h</td> | ||
| + | <td>7.32 </td> | ||
| + | <td>1.39</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>lycopene epsilon-cyclase</td> | ||
| + | <td>LCY</td> | ||
| + | <td>Oryza sativa cv.</td> | ||
| + | <td>44.66</td> | ||
| + | <td>200 nM/h</td> | ||
| + | <td>6.67 </td> | ||
| + | <td>1.27</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>carotene 3-hydroxylase</td> | ||
| + | <td>HYD</td> | ||
| + | <td>Oryza sativa cv.</td> | ||
| + | <td>53.70</td> | ||
| + | <td>200 nM/h</td> | ||
| + | <td>7.89 </td> | ||
| + | <td>1.5</td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <div class="p_font_size">However, the experimental data show great difference from our prediction model. Some genes are supported to be strong expressed in our model while few expressions were detected actually. Therefore, we believe that there are relationships between different genes we mentioned. For example, the gene PSY control the expression of the phytoene synthase, which is determinant transformation from pre-phytoene diphosphate to phytoene and phytoene is substrate of the following reaction, which was catalyzed by phytoene desaturase(encoded b gene CrtI/PDS).In this way, gene PSY and CrtI/PDS have strong relationships so the expression of one gene affect the other one.</div> | ||
<div class="row" style="margin-top:20px"> | <div class="row" style="margin-top:20px"> | ||
| − | <div class="col | + | <div class="col s12" align="center"> |
| − | + | <img alt="image" class="img-responsive col s11" src="https://static.igem.org/mediawiki/2016/d/d9/T--SCAU-China--model_p3.jpg"> | |
| − | + | ||
| − | + | ||
| − | <img alt="image" class="img-responsive col s11" src="https://static.igem.org/mediawiki/2016/ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="p_font_size" style="margin-bottom:20px"><small>Figure 12 qRT-PCR analyses of foreign and endogenous genes involved in carotenoids biosynthesis in aSTARice endosperm.</small></div> | |
| − | </div> | + | <div class="p_font_size"><small>The PDS, ZDS, ISO, BLCY, ELCY, HYD and rPSY ,all are rice endogenous genes for carotenoid biosynthesis.</small></div> |
| − | </div> | + | <div class="p_font_size">The data show that there is a positive correlation between the expression of CrtI gene and PSY gene in the indica transgenic rice HG1. The expression of BKT and BHY are fluctuant. Some transgenic lines have low expression of BKT and high expression of BHY while some strains have low expression of BHY but high expression of BKT and CrtI. All endogenous genes involved in carotenoid biosynthesis are low or no expression in aSTARice endosperm. The production and accumulation of astaxanthin in aSTRice is determined by co-expression of PSY, Crtl, BHY and BKT.</div> |
| + | <div class="p_font_size">Reference:</div> | ||
| + | <div class="p_font_size">【1】Alon U. An introduction to systems biology: design principles of biological circuits [M]. CRC press, 2006.</div> | ||
| + | <div class="p_font_size">【2】Ingalls B. Mathematical Modelling in Systems Biology: An Introduction[J]. Internet.[cited at p. 117], 2013.</div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
</body> | </body> | ||
Revision as of 07:32, 14 October 2016
Modeling
Astaxanthin is becoming a more and more popular health care product, and our project is focusing on producing and accumulating astaxanthin in rice endosperm bioreactor, and, finally, getting the astaxanthin products. During our experiments, we have collected some data and used educated guesses that are biologically feasible, trying to figure out, 1.which foreign genes plays a more important role in the pathway; 2. The interaction between four foreign genes and other endogenous genes on expression level. Through these analyses, we wonder a better optimizing strategy to increase the production of astaxanthin in rice endosperm bioreactor.
1. Genes and astaxanthin productiont
We have transferred four foreign genes, PSY, Crtl, BKT and BHY, into rice endosperm bioreactor. The enzymes encoded by these genes catalyze the following reactions:

Figure 10 The biosynthesis pathway of astaxanthin formation in transgenic rice endosperm. The dotted arrows indicate pathway limitations in rice endosperm. The solid arrows indicate the existence of carotenogenic reactions. The red arrows indicate the reactions catalaysed by four exogenous transgenes Psy, CrtI, BHY and BKT.

Figure 11 Correlation analyses between expression levels of four genes involved in astaxanthin biosynthesis and astaxanthin content.
The figure 11 showed us the correlations between astaxanthin concentration and expression of astaxanthin biosynthetic genes in rice endosperm bioreactor. OsActin1 gene was used to normalize expression. The encoded enzyme Crtl catalyzes the phytoene into lycopene, while enzyme BHY catalyzes the β-carotene into zeaxanthin. The expression of Crtl and BHY was significantly positively correlated with astaxanthin concentration in the rice endosperm bioreactor. The result suggests that the expression of Crtl and BHY are the rate-limiting factors to astaxanthin biosynthesis in rice endosperm bioreactor. Learning about this, in the future, we might increase the production of astaxanthin by enhancing the expression of Crtl and BHY.
2. Foreign genes and endogenous genesof
To synthesize astaxanthin in rice endosperm bioreactor, we have transduced four foreign genes, thus, we want to know the interplay between foreign genes and endogenous genes, or will their expression influence each other. Especially, we focus on the genes’ transcription network. The input signals usually change transcription factor activities on a sub-second timescale. Binding of the active transcription factor to its DNA sites often reaches equilibrium in seconds. Transcription and translation of the target gene takes minutes, and the accumulation of the protein can take minutes to hours.of
The production of the protein encoded by gene Y is balanced by two process, protein degradation( its specific destruction by specialized in the cell) and dilution( the reduction in concentration due to the increase of cell volume during growth). The degradation rate is , and the dilution rate is
, and the dilution rate is , giving a total degradation/dilution rate of
, giving a total degradation/dilution rate of
 , and the dilution rate is
, and the dilution rate is , giving a total degradation/dilution rate of
, giving a total degradation/dilution rate of
The change in concentration of protein Y is due to the difference between its production and degradation/dilution, as described by a dynamic equation:

At steady state, protein Y reaches a constant concentration . The steady-state concentration can be found by solving for
. The steady-state concentration can be found by solving for . This shows that the steady-state concentration is the ratio of the production and degradation/dilution rates:
. This shows that the steady-state concentration is the ratio of the production and degradation/dilution rates:
 . The steady-state concentration can be found by solving for
. The steady-state concentration can be found by solving for . This shows that the steady-state concentration is the ratio of the production and degradation/dilution rates:
. This shows that the steady-state concentration is the ratio of the production and degradation/dilution rates:
If we take away the input signal, so that production of protein Y stops and there will be an exponential decay of Y concentration:

Similarly, if an unstimulated cell with Y=0 is provided with a signal, so that protein Y begins to accumulate. If an unstimulated gene becomes suddenly stimulated by a strong signal Sx, the dynamic equation approach to steady state:

T1/2 is defined as the time to reach halfway between the initial and final levels in a dynamic process:
T1/2 =log(2)/α
T1/2 =log(2)/α

The concentration of Y rises from zero and gradually converges on the steady-state.
Note that at early times, when ,we can use a Taylor expansion to find a linear accumulation of a Y:
,we can use a Taylor expansion to find a linear accumulation of a Y:
 ,we can use a Taylor expansion to find a linear accumulation of a Y:
,we can use a Taylor expansion to find a linear accumulation of a Y:Y~βt13
early times, 

The concentration of protein Y accumulates at early times with a slope equal to its production rate. Later, as Y levels increase, the degradation term- Y begins to be important and Y converges to its steady-state level.
Y begins to be important and Y converges to its steady-state level.
 Y begins to be important and Y converges to its steady-state level.
Y begins to be important and Y converges to its steady-state level.We considered the activation of transcription of a gene( mRNA production) and used a dynamical equation to describe the changes in the concentration of the gene product, the protein Y. The mRNA needs to be translated to form the protein, and mRNA itself is also degraded by specific enzymes.
Assuming that mRNA is produced at rate and degraded at rate
and degraded at rate , and that each mRNA produces on average p protein molecules over its lifetime. The protein is degraded/diluted at rate
, and that each mRNA produces on average p protein molecules over its lifetime. The protein is degraded/diluted at rate .
.
 and degraded at rate
and degraded at rate , and that each mRNA produces on average p protein molecules over its lifetime. The protein is degraded/diluted at rate
, and that each mRNA produces on average p protein molecules over its lifetime. The protein is degraded/diluted at rate .
.The dynamic equation for the concentration of mRNA of gene Y, , is :
, is :
 , is :
, is : ①
①
The dynamical equation for the protein product is due to production of  copies per mRNA and degradation/dilution at rate :
copies per mRNA and degradation/dilution at rate :
 copies per mRNA and degradation/dilution at rate :
copies per mRNA and degradation/dilution at rate :

The steady-state mRNA level is found by setting in Equation ①, yielding
in Equation ①, yielding
 in Equation ①, yielding
in Equation ①, yielding
Using this for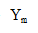 in Equation ① yields the following equation for the protein production rate:
in Equation ① yields the following equation for the protein production rate:
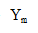 in Equation ① yields the following equation for the protein production rate:
in Equation ① yields the following equation for the protein production rate:
In other words, the effective protein production rate is equal to the steady-state mRNA level times the number of proteins translated from each mRNA:

In the first step of our modeling, we support that all genes have the same expression way in the cell. We support that all the input signals appear at time t=0. Because we don’t have enough time to measure transcription and translation rates of the genes, we assume that all the proteins have the same production rate and this rate are supported to be 200 nM/h. To measure the degradation rate of protein, αdeg, T1/2 is used in the following calculation, in which period of time the protein decrease by 50%.T1/2 of each protein is determined by their Instability index (II). Degradation rates of protein can be calculated by the aquation, T1/2 =log(2)/α.Yst means steady-state concentration of the proteins.
| Protein names | Gene names | Organism | Instability index (II) | β | αdeg | Yst |
|---|---|---|---|---|---|---|
| Actin | OsAct1 | Oryza sativa subsp. japonica | 34.96 | 200 nM/h | 5.26 | 1 |
| phytoene synthase | PSY | Zea mays L. | 59.82 | 200 nM/h | 9.09 | 1.73 |
| phytoene desaturase | CrtI/PDS | Erwinia uredovora | 31.38 | 200 nM/h | 4.69 | 0.89 |
| β-carotene hydroxylase | BHY | Haematococcus pluvialis | 52.32 | 200 nM/h | 7.89 | 1.50 |
| β-carotene ketolase | BKT | Chlamydomonas reinhardtii | 40.90 | 200 nM/h | 6.12 | 1.16 |
| phytoene desaturase | PDS | Oryza sativa cv. | 49.69 | 200 nM/h | 7.5 | 1.43 |
| ζ-carotene desaturase | ZDS | Oryza sativa cv. | 49.12 | 200 nM/h | 7.32 | 1.39 |
| lycopene epsilon-cyclase | LCY | Oryza sativa cv. | 44.66 | 200 nM/h | 6.67 | 1.27 |
| carotene 3-hydroxylase | HYD | Oryza sativa cv. | 53.70 | 200 nM/h | 7.89 | 1.5 |
However, the experimental data show great difference from our prediction model. Some genes are supported to be strong expressed in our model while few expressions were detected actually. Therefore, we believe that there are relationships between different genes we mentioned. For example, the gene PSY control the expression of the phytoene synthase, which is determinant transformation from pre-phytoene diphosphate to phytoene and phytoene is substrate of the following reaction, which was catalyzed by phytoene desaturase(encoded b gene CrtI/PDS).In this way, gene PSY and CrtI/PDS have strong relationships so the expression of one gene affect the other one.

Figure 12 qRT-PCR analyses of foreign and endogenous genes involved in carotenoids biosynthesis in aSTARice endosperm.
The PDS, ZDS, ISO, BLCY, ELCY, HYD and rPSY ,all are rice endogenous genes for carotenoid biosynthesis.
The data show that there is a positive correlation between the expression of CrtI gene and PSY gene in the indica transgenic rice HG1. The expression of BKT and BHY are fluctuant. Some transgenic lines have low expression of BKT and high expression of BHY while some strains have low expression of BHY but high expression of BKT and CrtI. All endogenous genes involved in carotenoid biosynthesis are low or no expression in aSTARice endosperm. The production and accumulation of astaxanthin in aSTRice is determined by co-expression of PSY, Crtl, BHY and BKT.
Reference:
【1】Alon U. An introduction to systems biology: design principles of biological circuits [M]. CRC press, 2006.
【2】Ingalls B. Mathematical Modelling in Systems Biology: An Introduction[J]. Internet.[cited at p. 117], 2013.

