| Line 16: | Line 16: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 1. The enzyme reaction by multiple complex<br>To connect different enzymes will<br>make continuous reaction efficiently. </h></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 32: | Line 32: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 2. Huge complex using SAP<br>To connect same enzymes like fluorescent proteins will amplify thir effects.</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 47: | Line 47: | ||
<div> | <div> | ||
<table class="merit"> | <table class="merit"> | ||
| − | < | + | <span class="small">Table. 1. Comparison between linkers and SAPs</span> |
<tr> | <tr> | ||
<th width="50%">Linker Method</th> | <th width="50%">Linker Method</th> | ||
| Line 89: | Line 89: | ||
<tr align="center" style="border-style: none"> | <tr align="center" style="border-style: none"> | ||
| − | <td style="border-style: none;">< | + | <td style="border-style: none;"><span class="small">Fig. 3. Using linkers<br>Expressions of gene A, B and C which code protein A, B and C are regulated by one promoter. If you connect some huge proteins, the expression efficiency may be decreased because the coding sequence is too long. </span></td> |
| − | <td style="border-style: none;">< | + | <td style="border-style: none;"><span class="small">Fig. 4. Using SAPs<br>You can produce protein A, B and C individually. After expression, they gather by SAPs and form disufide bonds by SLs.</span></td> |
</tr> | </tr> | ||
</center> | </center> | ||
| Line 107: | Line 107: | ||
<td style="border-style:none; float:center"><img src="https://static.igem.org/mediawiki/2016/2/23/T--HokkaidoU_Japan--multimerization_image3.png" alt="steric hindrance" height="300px" width="auto"> </td> </tr> | <td style="border-style:none; float:center"><img src="https://static.igem.org/mediawiki/2016/2/23/T--HokkaidoU_Japan--multimerization_image3.png" alt="steric hindrance" height="300px" width="auto"> </td> </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 5. Demerit of using linkers<br>In linker method, you need to consider the linker length to avoid the steric hindrance.</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 134: | Line 134: | ||
<tr align="center" style="border-style: none"> | <tr align="center" style="border-style: none"> | ||
| − | <td style="border-style: none;">< | + | <td style="border-style: none;"><span class="small">Fig. 6. Demerit of using SAP method<br>If some kinds of protein are expressed,<br>there are so many combination.<br>You may not be able to get the correct combination.</span></td> |
| − | <td style="border-style: none;">< | + | <td style="border-style: none;"><span class="small">Fig. 7. Resolution for infinite combinations<br>When you use some kinds of SAP,<br>incorrect connections will decrease.</span></td> |
</tr> | </tr> | ||
| Line 172: | Line 172: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 8. Design of the coding sequence</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 185: | Line 185: | ||
<tr><td style="border-style: none;"> | <tr><td style="border-style: none;"> | ||
<center><img src="https://static.igem.org/mediawiki/2016/0/0c/T--HokkaidoU_Japan--multimerization_image8.png" alt="methods" height="550px" width="auto"></center></td></tr> | <center><img src="https://static.igem.org/mediawiki/2016/0/0c/T--HokkaidoU_Japan--multimerization_image8.png" alt="methods" height="550px" width="auto"></center></td></tr> | ||
| − | <tr><td style="border-style: none;">< | + | <tr><td style="border-style: none;"><span class="small">Fig. 9. Method for verifying whether proteins form multiple complex </span></td></tr> |
</table> | </table> | ||
| Line 220: | Line 220: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 10. The construction of multimerization using SAP<br>This is the construct for making multiple complex. We used RADA16-I and P<span class="sitatuki">11</span>-4 as SAP. <br>C is a cysteine residues in short linker.</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 233: | Line 233: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 11. The construction of a negative control<br>We made a negative control which had only GFP to test the effect of SAPs.</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 247: | Line 247: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 12. Expected forming multiple complex</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 274: | Line 274: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center">< | + | <td style="border-style: none"; align="center"><span class="small">Fig. 13. The construction for making subunits of artificial multi-enzyme-complex<br>We designed this construct to had a cloning site. If you design the protein which ends are BamHI site, you can make the multimer easily.</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
Revision as of 11:21, 19 October 2016
Team:HokkaidoU Japan
\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\

We made a platform of technology for constructing covalently linked multi-enzyme-complex through disulfide bonds recruited by self-assembling peptide (SAP). By fusing SAP to the end of a protein, it will condense with other proteins’ SAP domains and form the complex. The SAP domains is pinched by short linkers (SL) that have cysteine residues. When the SAPs gather and SLs get close, disulfide bonds are formed between other SLs. So, we will make unbreakable complex. By using this method, we’ll be able to connect several enzymes and allow huge complexed proteins to be formed. It’ll improve the efficiency of a continuous reaction.
However, the ordinary method uses linkers to connect proteins. We think the new method using SAP is superior to the ordinary one for these reasons (Table. 1).
Table. 1. Comparison between linkers and SAPs
We thought the SAP method was best one but it had also disadvantages. Since the number of the possible combination of several different proteins is infinite, there is no guarantee that we can always obtain the expected combination.
One solution to the problem is limiting the number of combination by using different SAP. That can reduce probability of incorrect connection a little.
Multimerization is very useful. As forming protein complex with different functions, this multimer let us create more functional units. When same kinds of protein are used, it’ll be a large block and its function is expected to be enhanced.
We tried to establish novel uses of SAP in this yaer. We challenged multimerization using it and not only used it but also made firmly connection.

We tried forming multimers using the self-assembling peptide (SAP), P11-4 (QQRFEWEFEQQ) and RADA16-I (RADARADARADARADA). And to make firmly bonds we designed short linker (GGCGG) called SL for short. We Connected SL and SAP to both ends of the protein. In this experiment, we used GFP as test (Fif. 8).
GFP’s molecular mass is 26891Da. When fusing with P11-4, it’s 31709Da. With RADA16-I, it’s 31943Da. When they form multimer, the molecular mass will be more than 60kDa. Consequently, we used the filter which filters out the proteins with mass of more than 50KDa.
For the evaluation, we ordered IDT the designed constructions and put them on the vectors. Then, we introduced them to E.coli. Using IPTG induction , the proteins were expressed. Causing bacteriolysis with freeze-thaw, we acquired the supernatant contains the proteins by centrifugal separation. Purifying the protein with Ni-affinity chromatography, we filtrated the solution to separate the proteins with mass of less than 50KDa. We irradiated 480nm light to filtrate and observed whether 580nm wave-length light was emitted.

We put above CDS (Fig. 8) into pET15b and expressed (Fig. 10). As negative control we made a construction containing GFP without SAPs and SLs (Fig. 11). GFPs with SAPs and SLs was expected to become multiple complexs (Fig. 12).

As future work, anyone can make multi-enzyme-complex if the protein is designed to have BamHI restriction enzyme sites in both ends. Our construction have also BamHI site at GEP ends. So, you can cut out the GFP and put on any protein using cloning site (Fig. 13).
[1] Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242?251.
[2] Castellana M1, Wilson MZ2, Xu Y3, Joshi P2, Cristea IM2, Rabinowitz JD4, Gitai Z2, Wingreen NS3. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014 Oct;32(10):1011-8.

 |
| Fig. 1. The enzyme reaction by multiple complex To connect different enzymes will make continuous reaction efficiently. |
We made a platform of technology for constructing covalently linked multi-enzyme-complex through disulfide bonds recruited by self-assembling peptide (SAP). By fusing SAP to the end of a protein, it will condense with other proteins’ SAP domains and form the complex. The SAP domains is pinched by short linkers (SL) that have cysteine residues. When the SAPs gather and SLs get close, disulfide bonds are formed between other SLs. So, we will make unbreakable complex. By using this method, we’ll be able to connect several enzymes and allow huge complexed proteins to be formed. It’ll improve the efficiency of a continuous reaction.
 |
| Fig. 2. Huge complex using SAP To connect same enzymes like fluorescent proteins will amplify thir effects. |
However, the ordinary method uses linkers to connect proteins. We think the new method using SAP is superior to the ordinary one for these reasons (Table. 1).
| Linker Method | SAP Method |
|---|---|
| Regulated by one promoter (Fig. 3) | Each protein can be produced individually (Fig. 4) |
| Difficult to produce several huge complex | Possible to synthesize the proteins individually. Can also form a huge complex (Fig. 4) |
| The possibility of deformation of the 3D-structure (Fig. 5) | Low possibility of deformation since they only connect with proteins which can condense |

|

|
| Fig. 3. Using linkers Expressions of gene A, B and C which code protein A, B and C are regulated by one promoter. If you connect some huge proteins, the expression efficiency may be decreased because the coding sequence is too long. |
Fig. 4. Using SAPs You can produce protein A, B and C individually. After expression, they gather by SAPs and form disufide bonds by SLs. |
 |
| Fig. 5. Demerit of using linkers In linker method, you need to consider the linker length to avoid the steric hindrance. |
We thought the SAP method was best one but it had also disadvantages. Since the number of the possible combination of several different proteins is infinite, there is no guarantee that we can always obtain the expected combination.
One solution to the problem is limiting the number of combination by using different SAP. That can reduce probability of incorrect connection a little.
 |

|
| Fig. 6. Demerit of using SAP method If some kinds of protein are expressed, there are so many combination. You may not be able to get the correct combination. |
Fig. 7. Resolution for infinite combinations When you use some kinds of SAP, incorrect connections will decrease. |
Multimerization is very useful. As forming protein complex with different functions, this multimer let us create more functional units. When same kinds of protein are used, it’ll be a large block and its function is expected to be enhanced.
We tried to establish novel uses of SAP in this yaer. We challenged multimerization using it and not only used it but also made firmly connection.

We tried forming multimers using the self-assembling peptide (SAP), P11-4 (QQRFEWEFEQQ) and RADA16-I (RADARADARADARADA). And to make firmly bonds we designed short linker (GGCGG) called SL for short. We Connected SL and SAP to both ends of the protein. In this experiment, we used GFP as test (Fif. 8).
 |
| Fig. 8. Design of the coding sequence |
Assay
 |
| Fig. 9. Method for verifying whether proteins form multiple complex |
GFP’s molecular mass is 26891Da. When fusing with P11-4, it’s 31709Da. With RADA16-I, it’s 31943Da. When they form multimer, the molecular mass will be more than 60kDa. Consequently, we used the filter which filters out the proteins with mass of more than 50KDa.
For the evaluation, we ordered IDT the designed constructions and put them on the vectors. Then, we introduced them to E.coli. Using IPTG induction , the proteins were expressed. Causing bacteriolysis with freeze-thaw, we acquired the supernatant contains the proteins by centrifugal separation. Purifying the protein with Ni-affinity chromatography, we filtrated the solution to separate the proteins with mass of less than 50KDa. We irradiated 480nm light to filtrate and observed whether 580nm wave-length light was emitted.

We put above CDS (Fig. 8) into pET15b and expressed (Fig. 10). As negative control we made a construction containing GFP without SAPs and SLs (Fig. 11). GFPs with SAPs and SLs was expected to become multiple complexs (Fig. 12).
 |
| Fig. 10. The construction of multimerization using SAP This is the construct for making multiple complex. We used RADA16-I and P11-4 as SAP. C is a cysteine residues in short linker. |
 |
| Fig. 11. The construction of a negative control We made a negative control which had only GFP to test the effect of SAPs. |
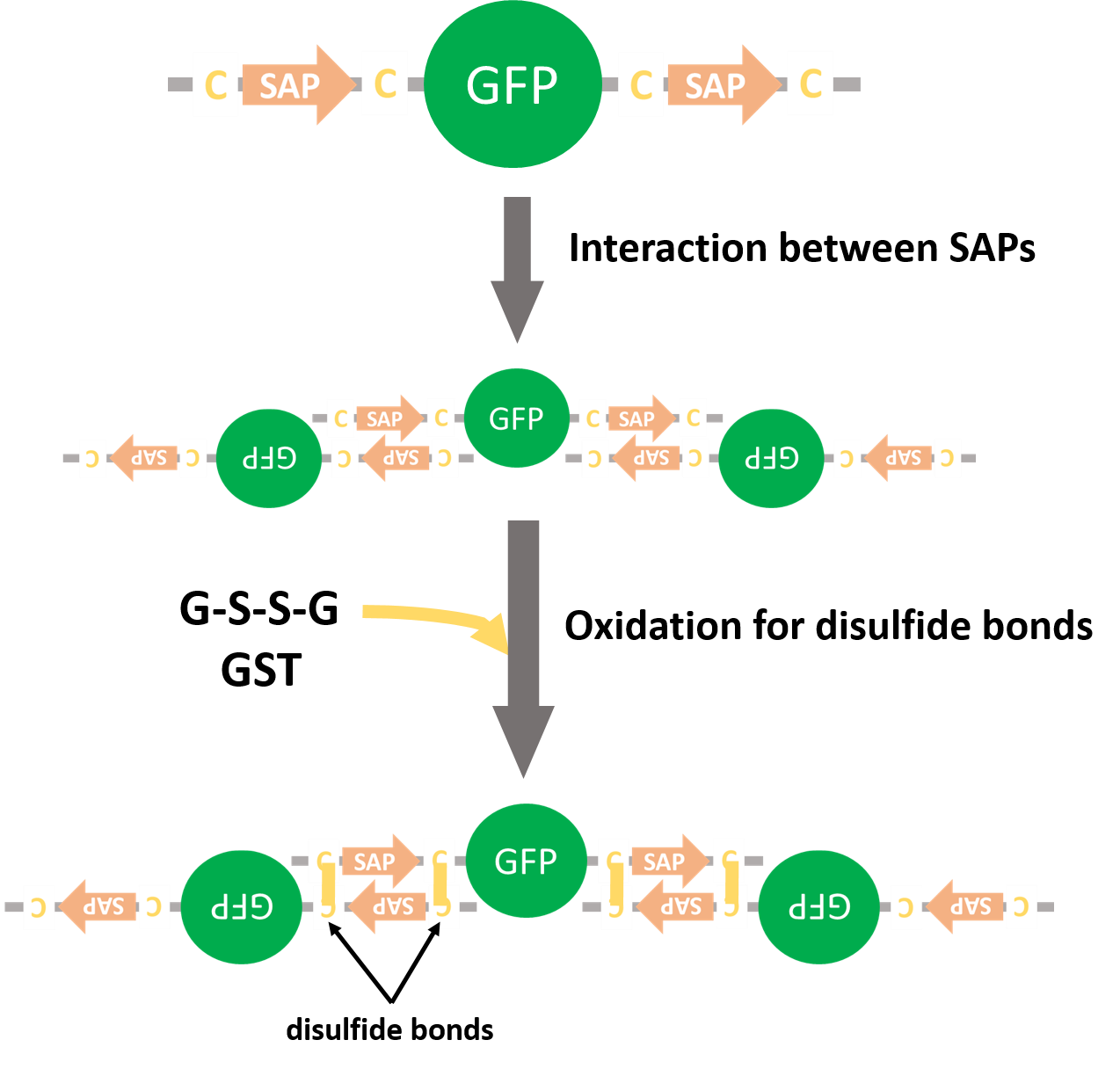 |
| Fig. 12. Expected forming multiple complex |

As future work, anyone can make multi-enzyme-complex if the protein is designed to have BamHI restriction enzyme sites in both ends. Our construction have also BamHI site at GEP ends. So, you can cut out the GFP and put on any protein using cloning site (Fig. 13).
 |
| Fig. 13. The construction for making subunits of artificial multi-enzyme-complex We designed this construct to had a cloning site. If you design the protein which ends are BamHI site, you can make the multimer easily. |
[1] Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242?251.
[2] Castellana M1, Wilson MZ2, Xu Y3, Joshi P2, Cristea IM2, Rabinowitz JD4, Gitai Z2, Wingreen NS3. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014 Oct;32(10):1011-8.

 "
"





