| (5 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
{{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/9/97/T--UBonn_HBRS--Enzyme_Assays_intro.jpg|alt=Intro Image|class=100}} | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/9/97/T--UBonn_HBRS--Enzyme_Assays_intro.jpg|alt=Intro Image|class=100}} | ||
| − | '''In order to | + | '''The efficiency of Enzymatic Deinking is directly dependent on the specific activity of the employed enzymes. In order to keep the the conditions constant between experiments while optimizing the deinking procedure, we developed specific enzymatic activity assays for our enzyme classes Endoglucanases and Xylanases.''' |
== Introduction == | == Introduction == | ||
The aim of the enzyme assays, is to determine the amount of enzymes present in the sample under | The aim of the enzyme assays, is to determine the amount of enzymes present in the sample under | ||
| − | certain conditions, making activity comparable between | + | certain conditions, making activity comparable between different samples and experiments. The activity |
| − | is described by the product formed over | + | is either described by the amount of product formed over a given time or the amount of substrate having been consumed. |
| − | Different parameters affecting the activity of | + | Different parameters affecting the activity of enzymes are substrates and their concentration, pH, |
ionic strength, buffer type and temperature. | ionic strength, buffer type and temperature. | ||
| − | In our project, | + | In our project, we employed stopped colorimetric assays. This means that the assays were |
| − | stopped after a fixed time | + | stopped after a fixed time by heat-inactivating the enzymes and the amount of product formed was |
| − | measured. Colorimetric | + | measured. Colorimetric assays makes it possible to easily quantify the reaction product by spectrophotometry, |
| − | + | often by converting it again with another reagent. The absorbance at a given wavelength is directly | |
| − | proportional to the product in the sample. In this experiment, 3,5-Dinitrosalicylic acid was used. | + | proportional to the amount of product in the sample. In this experiment, 3,5-Dinitrosalicylic acid was used. |
| − | The enzymes tested were | + | The enzymes tested were cellulases, specfically endoglucanases (endo-1,4-β-D-glucanase, ''Acidothermus cellulolyticus'', |
| − | Sigma-Aldrich) and | + | Sigma-Aldrich) and xylanases (endo-1,4-β-Xylanase M4, ''Aspergillus niger'', Megazyme). The fromer |
| − | + | breaks down cellulose molecules by hydrolysis of the 1,4-beta-D- | |
glycosidic linkages into monosaccahrides such as beta-glucose. Xylanase breaks down | glycosidic linkages into monosaccahrides such as beta-glucose. Xylanase breaks down | ||
hemicellulose by degrading polysaccahride beta-1,4-xylan (Beechwood) into xylose. | hemicellulose by degrading polysaccahride beta-1,4-xylan (Beechwood) into xylose. | ||
| − | == Experimental | + | == Experimental Setup and Validation == |
| − | The tests done always follow similar recipe (see protocol) that | + | The tests done always follow a similar recipe (see protocol) that contains sugar or enzyme solution, |
citrate buffer, water, substrate and DNS. | citrate buffer, water, substrate and DNS. | ||
Buffers are needed to adjust and stabilize the pH for enzyme assays. The concentration that works | Buffers are needed to adjust and stabilize the pH for enzyme assays. The concentration that works | ||
for our chosen enzymes is 50mM. | for our chosen enzymes is 50mM. | ||
| − | Substrate used for endoglucanase was Carboxymethyl cellulose (CMC) which is a cellulose | + | The Substrate used for endoglucanase was Carboxymethyl cellulose (CMC) which is a cellulose |
derivative that the enzyme is able to cleave. Initially for the standard calibrations 1:3 CMC was | derivative that the enzyme is able to cleave. Initially for the standard calibrations 1:3 CMC was | ||
used but in order to increase the availability of the substrate, the assays with enzymes had CMC 3:7. | used but in order to increase the availability of the substrate, the assays with enzymes had CMC 3:7. | ||
| Line 37: | Line 37: | ||
DNS is aromatic compound that reacts with reducing sugars to form 3-amino-5-nitrosalicylic acid- | DNS is aromatic compound that reacts with reducing sugars to form 3-amino-5-nitrosalicylic acid- | ||
| − | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/6/6a/T--UBonn_HBRS--Enzyme_Assays_Kyoto.jpg|alt=Kyoto digestion DNS assay|class=100|caption-class=with-caption|caption=<i> | + | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/6/6a/T--UBonn_HBRS--Enzyme_Assays_Kyoto.jpg|alt=Kyoto digestion DNS assay|class=100|caption-class=with-caption|caption=<i>DNS digest assay</i>}} |
This absorbs light at 540 nm creating a possibility to quantify the product present in the sample. | This absorbs light at 540 nm creating a possibility to quantify the product present in the sample. | ||
| Line 125: | Line 125: | ||
The results of the endoglucanase assays are illustrated on the upcoming graphs. | The results of the endoglucanase assays are illustrated on the upcoming graphs. | ||
| + | |||
| + | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/4/40/T--UBonn_HBRS--Enzyme_Assays_glucose_calibration2.png|alt=Glucose Calibration Endoglucanase|class=100|caption-class=with-caption|caption=<strong>Figure 7. Standard calibration of the Glucose for Endoglucanase.</strong> Absorbance is put against the glucose concentrations in μmol.}} | ||
| + | |||
| + | |||
| + | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/7/7a/T--UBonn_HBRS--Enzyme_Assays_endoglucanase_assay.png|alt=Glucose Calibration Endoglucanase|class=100|caption-class=with-caption|caption=<strong>Figure 8. Endoglucanase Assay.</strong> Absorbance of the samples shown in relationship to corresponding enzyme Units.}} | ||
| + | |||
| + | |||
| + | The graphs show linear ranges but error bars can be considered big. This is especially true for the | ||
| + | enzyme experiment. Units used went up to 0,25 U and absorbance maxima was 0,225. Going over | ||
| + | the amount of enzymes produced saturated curve. Lower values also showed results that can not be | ||
| + | take into account, as they showed random results opposed to constant increase with increasing | ||
| + | concentrations. | ||
| + | |||
| + | |||
| + | === Xylanase Assay === | ||
| + | Xylanase assays were successful and the results can be seen on the graphs below. Standards were | ||
| + | done in parallel to the enzyme assay. | ||
| + | |||
| + | |||
| + | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/d/d2/T--UBonn_HBRS--Enzyme_Assays_xylose_calibration2.png|alt=Xylose Calibration |class=100|caption-class=with-caption|caption=<strong>Figure 9. Standard calibration of the Xylose for Xylanase Assay.</strong> Absorbance results are plotted against sugar concentration in μmol.}} | ||
| + | |||
| + | |||
| + | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/c/ce/T--UBonn_HBRS--Enzyme_Assays_xylanase_assay.png|alt=Xylanase Assay|class=100|caption-class=with-caption|caption=<strong>Figure 10. Xylanase Assay.</strong> Absorbance in reference to the enzyme units used in the sample.}} | ||
| + | |||
| + | |||
| + | Both graphs show a linear trend. Highest amount of units used was 0,1U, which resulted in an absorbance around 0,75. | ||
| + | |||
| + | |||
| + | ==== Discussion ==== | ||
| + | The experiments done with the enzymes gave analysable results. The proper linear | ||
| + | trend line of standard calibration shows that the the samples were treated properly | ||
| + | and acted as in experiments done before. | ||
| + | |||
| + | Endoglucanase assays show large deviations but give a linear trend line. Deviations | ||
| + | source is mainly the properties of solutions used in the experiment and reasons that | ||
| + | will be discussed in next sub-chapter. | ||
| + | |||
| + | The enzyme assay gives lower absorbance results which is likely due to the properties | ||
| + | of the enzyme and substrate. Endoglucanase has lower activity to begin with in | ||
| + | comparison to Xylanase. It could also be that the amount of substrate present was not | ||
| + | enough to show full activity. However, tests were done in different CMC | ||
| + | concentrations to find the point where the trend line becomes saturated. | ||
| + | |||
| + | In case of xylanase, the results show a expected linear patter and deviations are low. | ||
| + | This can be contrasted to viscosity of substrate for endoglucanase. Xylan is much | ||
| + | more easy to pipet, not leading to as many differences that could come from residual | ||
| + | liquids. It also shows higher activity based on information provided my manufacturer. | ||
| + | |||
| + | All in all, the experiments were successful but improvable. | ||
| + | |||
| + | |||
| + | ==== Troubleshooting and Missing Parts ==== | ||
| + | Probable sources of errors are pipetting, time differences in | ||
| + | preparations of the samples, room temperature, decay of enzymatic activity and | ||
| + | viscosity of solutions. Measures must be taken where possible, in order to decrease | ||
| + | the amount of deviations. | ||
| + | |||
| + | |||
| + | ==== Protocols ==== | ||
| + | All experiments followed same principle and similar recipe. | ||
| + | |||
| + | The recipe for the experiments can be seen in the following table. | ||
| + | |||
| + | |||
| + | {{UBonn_HBRS/table-start|class=with-caption}} | ||
| + | <html> | ||
| + | <thead> | ||
| + | <tr> | ||
| + | <th></th> | ||
| + | <th></th> | ||
| + | <th>µl</th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td>1.</td> | ||
| + | <td><strong>Citrate (50 mM)</strong></td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2.</td> | ||
| + | <td><strong>Sugar/enzyme</strong></td> | ||
| + | <td>10</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>3.</td> | ||
| + | <td><strong>CMC /Beechwood</strong></td> | ||
| + | <td>75</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>4.</td> | ||
| + | <td><strong>ddH2O/Glycerol/Citrate</strong></td> | ||
| + | <td>10</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td><strong>Total Volume (without dns)</strong></td> | ||
| + | <td><strong>100</strong></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5.</td> | ||
| + | <td><strong>DNS</strong></td> | ||
| + | <td>100</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td><strong>Total Volume</strong></td> | ||
| + | <td><strong>200</strong></td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </html> | ||
| + | {{UBonn_HBRS/table-end}} | ||
| + | <html><div class="caption"></html> | ||
| + | <strong>Table 1. Enzyme Assay recipe.</strong> | ||
| + | <html></div></html> | ||
| + | |||
| + | |||
| + | 1. Citrate buffer: to be added in every experiment; 2. Sugar solution for standard calibration and | ||
| + | enzyme solutions for enzyme assays; 3. CMC for endoglucanase assays and glucose assays, | ||
| + | Beechwood for xylanase and xylose assays; 4. Glycerol and Citrate for its corresponding assays or | ||
| + | water as substitute where not used; 5. DNS to be used in all experiments done. | ||
| + | |||
| + | Citrate buffer, sugar or enzyme, substrate and water, glycerol or additional citrate depending on the | ||
| + | assay in question, are to be mixed according to the table given in the 200 μl PCR strips. Centrifuge | ||
| + | the droplets down. <br /> | ||
| + | This is followed by the first incubation: 50 oC for 30 minutes. The process is stopped by cooling the | ||
| + | samples down, for example, by using PCR cooler rack. | ||
| + | |||
| + | DNS is added to the samples, mixed and centrifuged and then incubated at 100 oC for 5 minutes. | ||
| + | The samples are cooled down again. | ||
| + | |||
| + | All the samples are transferred to 96 well plate in volume of 100 μl and scanned with | ||
| + | spectrophotometer in 540 nm and 750 nm. | ||
{{UBonn_HBRS/footer}} | {{UBonn_HBRS/footer}} | ||
Latest revision as of 23:36, 19 October 2016
Enzyme Assays

The efficiency of Enzymatic Deinking is directly dependent on the specific activity of the employed enzymes. In order to keep the the conditions constant between experiments while optimizing the deinking procedure, we developed specific enzymatic activity assays for our enzyme classes Endoglucanases and Xylanases.
Contents
Introduction
The aim of the enzyme assays, is to determine the amount of enzymes present in the sample under certain conditions, making activity comparable between different samples and experiments. The activity is either described by the amount of product formed over a given time or the amount of substrate having been consumed.
Different parameters affecting the activity of enzymes are substrates and their concentration, pH, ionic strength, buffer type and temperature.
In our project, we employed stopped colorimetric assays. This means that the assays were stopped after a fixed time by heat-inactivating the enzymes and the amount of product formed was measured. Colorimetric assays makes it possible to easily quantify the reaction product by spectrophotometry, often by converting it again with another reagent. The absorbance at a given wavelength is directly proportional to the amount of product in the sample. In this experiment, 3,5-Dinitrosalicylic acid was used.
The enzymes tested were cellulases, specfically endoglucanases (endo-1,4-β-D-glucanase, Acidothermus cellulolyticus, Sigma-Aldrich) and xylanases (endo-1,4-β-Xylanase M4, Aspergillus niger, Megazyme). The fromer breaks down cellulose molecules by hydrolysis of the 1,4-beta-D- glycosidic linkages into monosaccahrides such as beta-glucose. Xylanase breaks down hemicellulose by degrading polysaccahride beta-1,4-xylan (Beechwood) into xylose.
Experimental Setup and Validation
The tests done always follow a similar recipe (see protocol) that contains sugar or enzyme solution, citrate buffer, water, substrate and DNS. Buffers are needed to adjust and stabilize the pH for enzyme assays. The concentration that works for our chosen enzymes is 50mM. The Substrate used for endoglucanase was Carboxymethyl cellulose (CMC) which is a cellulose derivative that the enzyme is able to cleave. Initially for the standard calibrations 1:3 CMC was used but in order to increase the availability of the substrate, the assays with enzymes had CMC 3:7. Substrate for xylanase was 1% xylan, also known as beechwood. DNS is aromatic compound that reacts with reducing sugars to form 3-amino-5-nitrosalicylic acid-
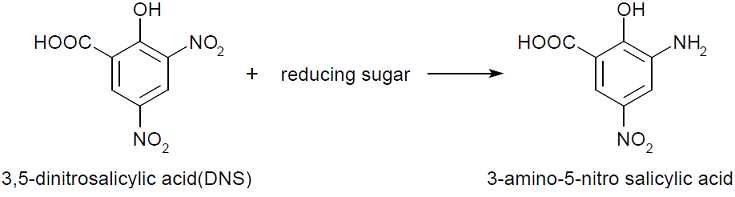
This absorbs light at 540 nm creating a possibility to quantify the product present in the sample.
Additionally, absorbance at 750 nm was measured in order to subtract the background absorbance
for normalization.
Additional water served purpose for having room to change different part of the recipe while still
keeping constant volume.
Analysis of the data was done with Open Office Calc.
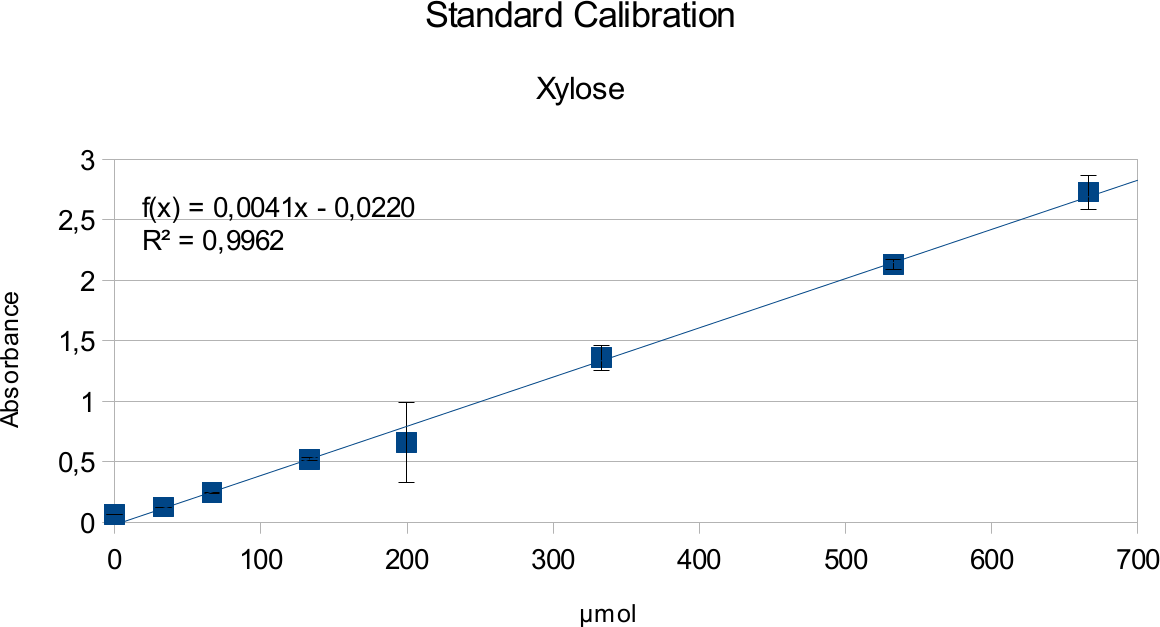
Standard Calibration
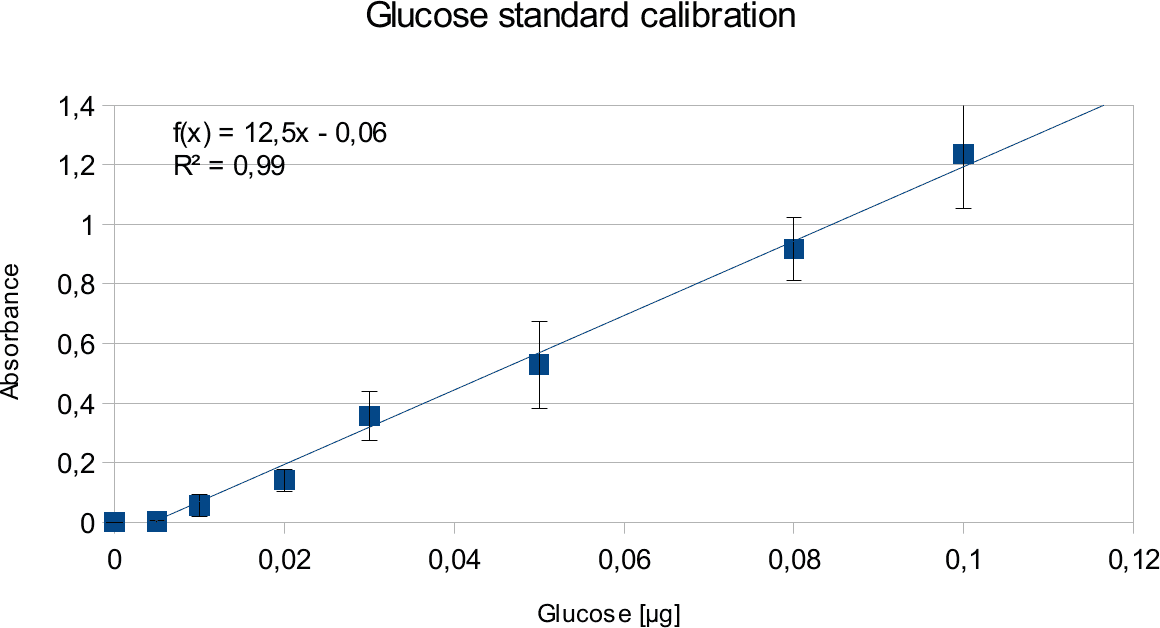
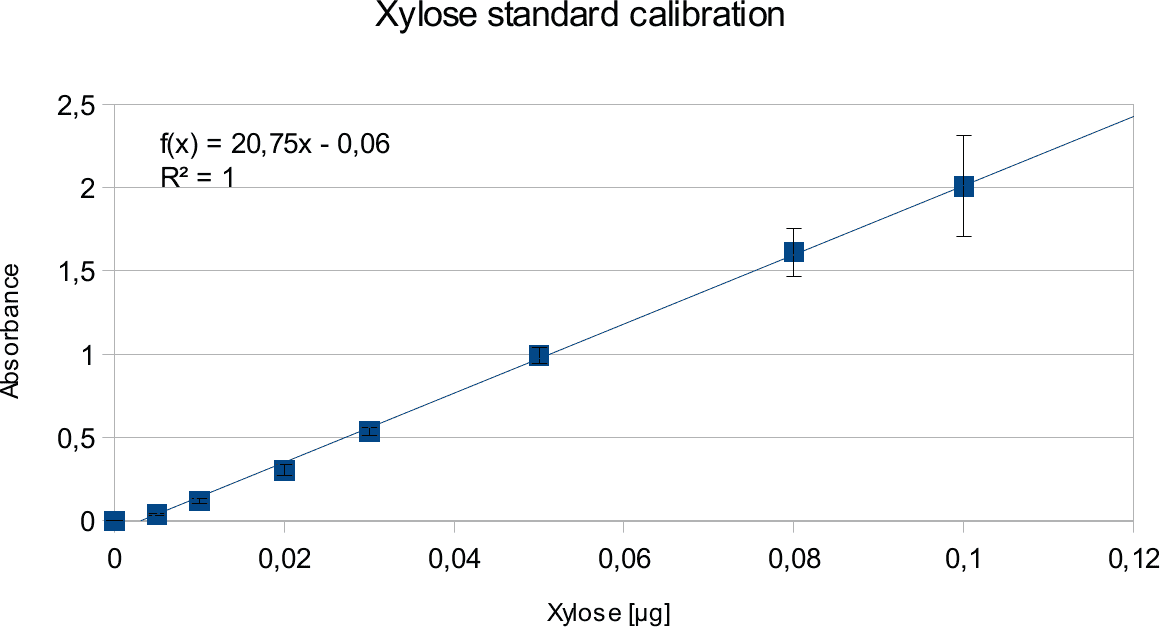
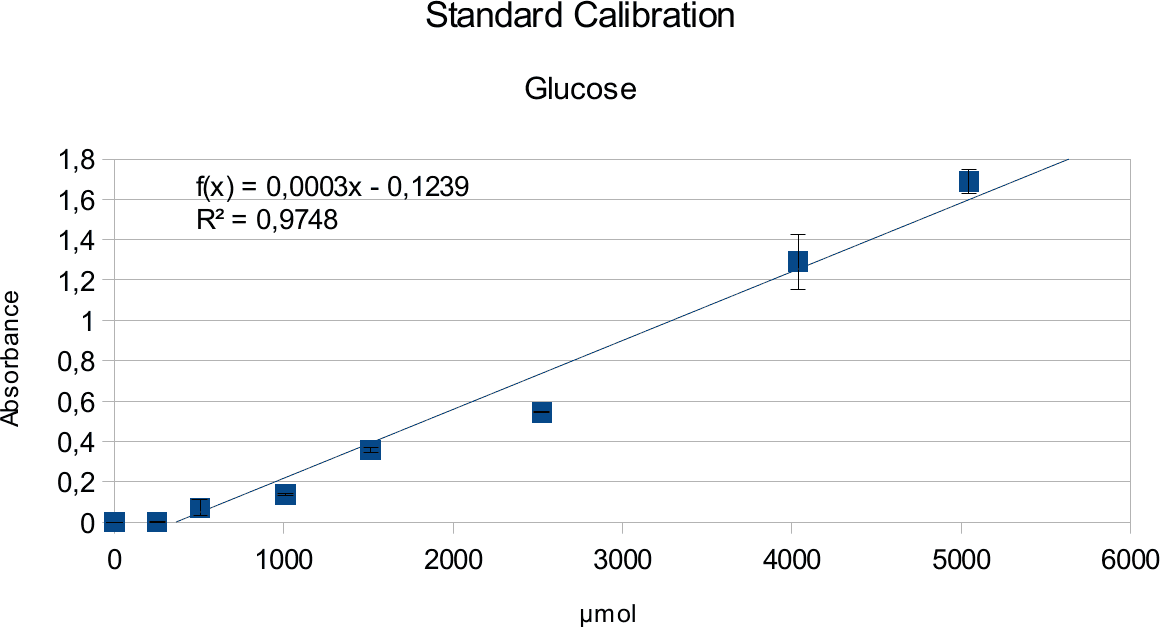
In order to determine the concentration of the substance of interest, standard calibration of the sample with known concentrations is necessary. Xylose was used for xylanase and glucose for endoglucanase.
Initially, a wide range of sugar concentration was tested which was 100 mg/ml to 0,001 mg/ml.
Through several experiments, a linear working range was determined to be 10 mg/ml to 0,5 mg/ml
(0,1to 0,005 μg in total in the sample)
Experiments creating a linear standard calibration lead to following results.
Discussion
The final results show linear range as expected and needed in order to validate the complete assay. Deviations increase with increasing concentration as smaller mistakes have then more effect. However, both xylose and glucose yielded usable results that aided in determination of the unit range, that should be used in experiments with enzymes.
Citrate and Glycerol Tests
In order to validate that the read out is not falsified by the variables in the test tube, a series of experiments were done. This entailed doing experiments with citrate buffer and glycerol. Buffer is needed to to create pH optimum and glycerol is enzyme stabilizing agent. Different concentrations of citrate buffer, glycerol and sugars were used.
A range of sugar concentrations of 0,1 μg to 0,0005 μg of total amount of sugar in sample was chosen for both experiments with exception of glucose citrate assay. The reason for higher high end glucose concentration, is due the fact that, results above 0,1 μg went over the measurable absorbance for other experiments and data point could not have been included. For citrate buffer, the concentrations were 1M to 0 M. In case of glycerol, the range was 90-0 %. The experimental set up followed same principle as before. The results can be viewed below.
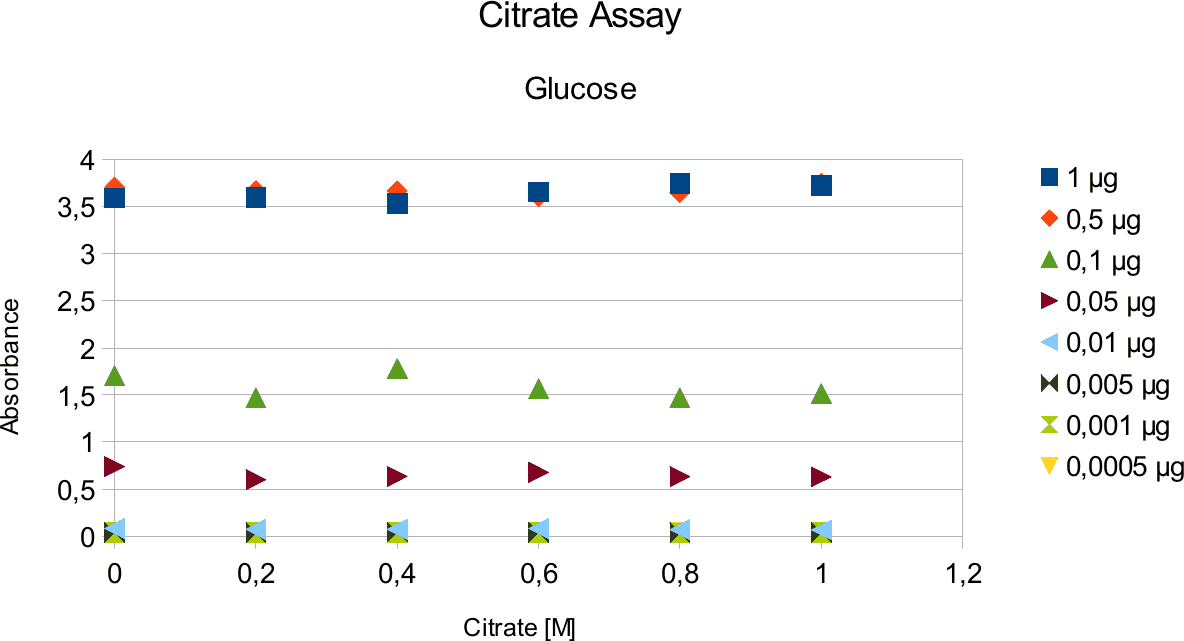
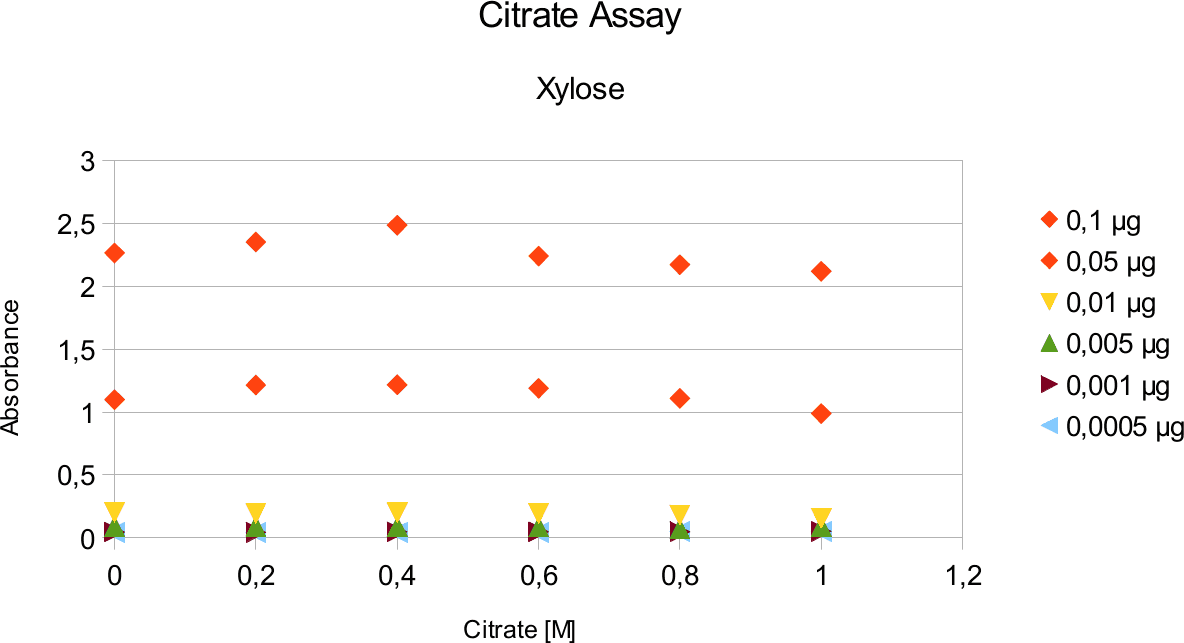
The obtained results show that the absorbance measured at was nearly constant for both, citrate
buffer assays with very low fluctuation.
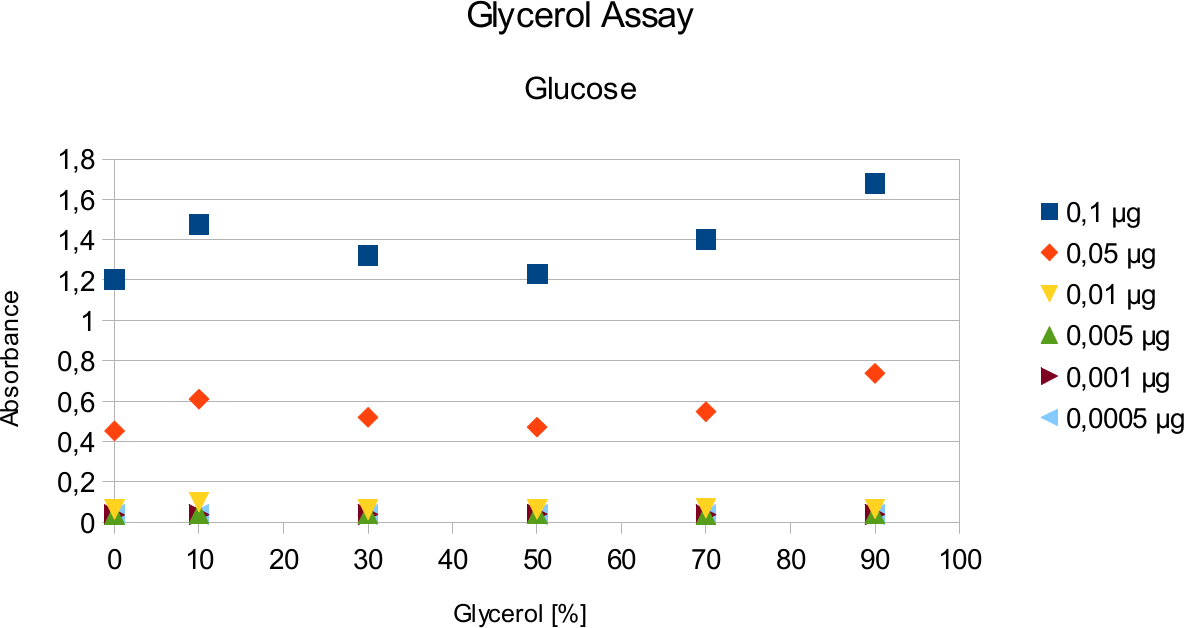
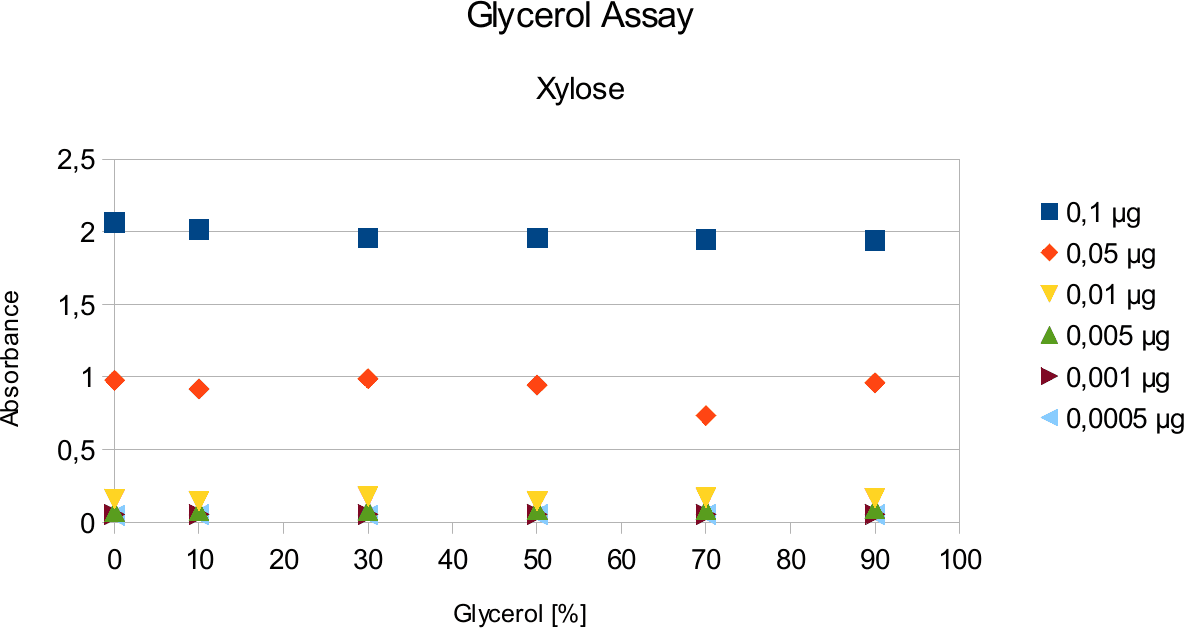
The results are similar to the ones from citrate. Here once again the absorbance values stay near
constant through all measurements.
Discussion
Citrate and glycerol result showed both similar pattern. There was some fluctuation between the samples, however, this can be taken as an error. Glycerol is rather viscous and therefore, difficult to pipette. Furthermore, sources of mistakes must be considered such as errors from pipetting. All in all, it can be concluded that citrate and glycerol have no effect in the experiments done.
Enzyme Assays
The assays done with xylanase and endoglucanase followed similar principle to the previous experiments. Precautions were taken to keep samples on cooler rack while preparing enzyme samples, in order to preserve the enzyme activity.
Endoglucanase Assays
Endoglucanase assays are made difficult by the substrate viscosity. This is the main source of deviations in these assays. Standard calibration was done in parallel to the enzyme experiments.
The results of the endoglucanase assays are illustrated on the upcoming graphs.
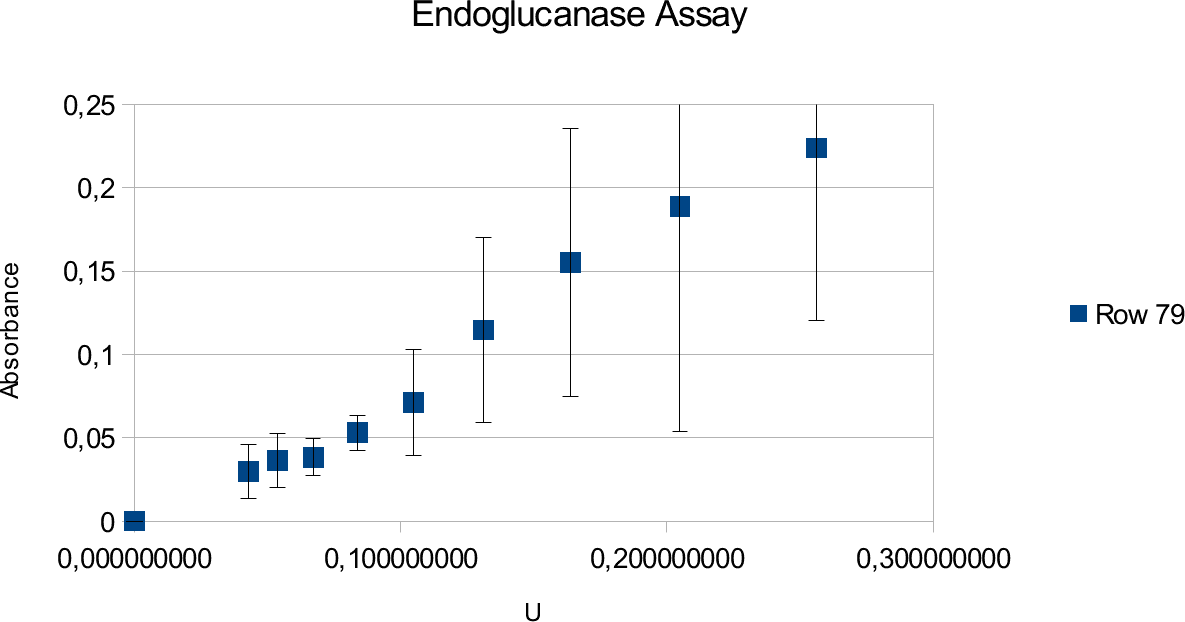
The graphs show linear ranges but error bars can be considered big. This is especially true for the
enzyme experiment. Units used went up to 0,25 U and absorbance maxima was 0,225. Going over
the amount of enzymes produced saturated curve. Lower values also showed results that can not be
take into account, as they showed random results opposed to constant increase with increasing
concentrations.
Xylanase Assay
Xylanase assays were successful and the results can be seen on the graphs below. Standards were done in parallel to the enzyme assay.
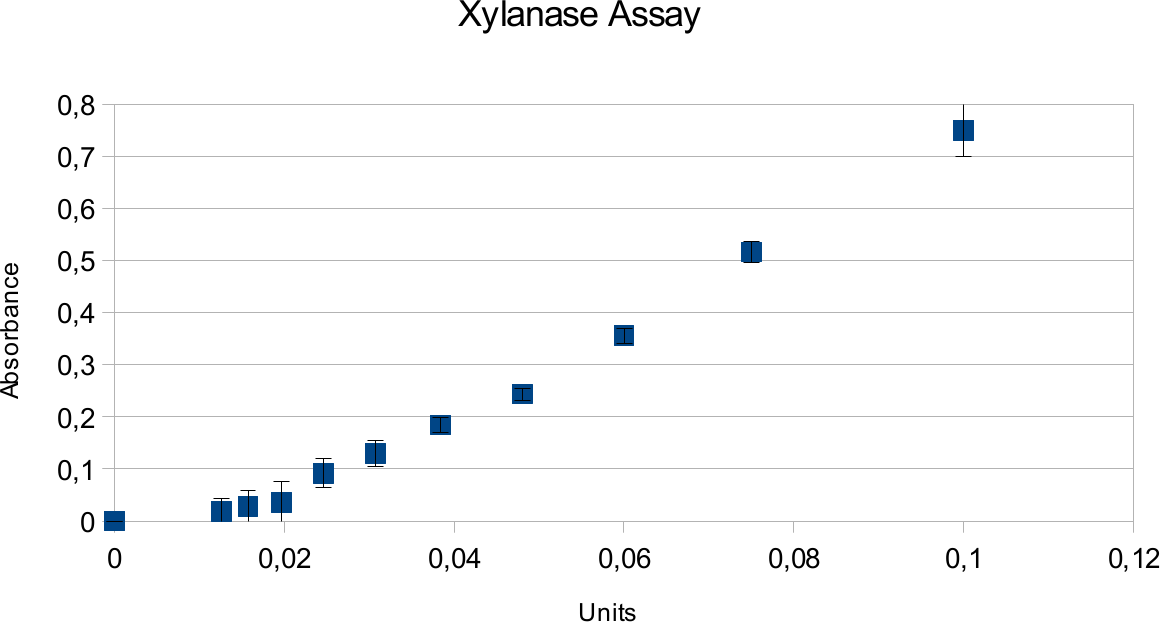
Both graphs show a linear trend. Highest amount of units used was 0,1U, which resulted in an absorbance around 0,75.
Discussion
The experiments done with the enzymes gave analysable results. The proper linear trend line of standard calibration shows that the the samples were treated properly and acted as in experiments done before.
Endoglucanase assays show large deviations but give a linear trend line. Deviations source is mainly the properties of solutions used in the experiment and reasons that will be discussed in next sub-chapter.
The enzyme assay gives lower absorbance results which is likely due to the properties of the enzyme and substrate. Endoglucanase has lower activity to begin with in comparison to Xylanase. It could also be that the amount of substrate present was not enough to show full activity. However, tests were done in different CMC concentrations to find the point where the trend line becomes saturated.
In case of xylanase, the results show a expected linear patter and deviations are low. This can be contrasted to viscosity of substrate for endoglucanase. Xylan is much more easy to pipet, not leading to as many differences that could come from residual liquids. It also shows higher activity based on information provided my manufacturer.
All in all, the experiments were successful but improvable.
Troubleshooting and Missing Parts
Probable sources of errors are pipetting, time differences in preparations of the samples, room temperature, decay of enzymatic activity and viscosity of solutions. Measures must be taken where possible, in order to decrease the amount of deviations.
Protocols
All experiments followed same principle and similar recipe.
The recipe for the experiments can be seen in the following table.
| µl | ||
|---|---|---|
| 1. | Citrate (50 mM) | 5 |
| 2. | Sugar/enzyme | 10 |
| 3. | CMC /Beechwood | 75 |
| 4. | ddH2O/Glycerol/Citrate | 10 |
| Total Volume (without dns) | 100 | |
| 5. | DNS | 100 |
| Total Volume | 200 |
1. Citrate buffer: to be added in every experiment; 2. Sugar solution for standard calibration and
enzyme solutions for enzyme assays; 3. CMC for endoglucanase assays and glucose assays,
Beechwood for xylanase and xylose assays; 4. Glycerol and Citrate for its corresponding assays or
water as substitute where not used; 5. DNS to be used in all experiments done.
Citrate buffer, sugar or enzyme, substrate and water, glycerol or additional citrate depending on the
assay in question, are to be mixed according to the table given in the 200 μl PCR strips. Centrifuge
the droplets down.
This is followed by the first incubation: 50 oC for 30 minutes. The process is stopped by cooling the
samples down, for example, by using PCR cooler rack.
DNS is added to the samples, mixed and centrifuged and then incubated at 100 oC for 5 minutes. The samples are cooled down again.
All the samples are transferred to 96 well plate in volume of 100 μl and scanned with spectrophotometer in 540 nm and 750 nm.


