Ayanchromium (Talk | contribs) |
Prabaha007 (Talk | contribs) |
||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 62: | Line 62: | ||
<div class="container-fluid"> | <div class="container-fluid"> | ||
<div class="navbar-header"> | <div class="navbar-header"> | ||
| − | + | </div> | |
<div class="collapse navbar-collapse" id="myNavbar"> | <div class="collapse navbar-collapse" id="myNavbar"> | ||
<ul class="nav navbar-nav"> | <ul class="nav navbar-nav"> | ||
<li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore">Home</a></li> | <li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore">Home</a></li> | ||
<li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Team">Team</a></li> | <li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Team">Team</a></li> | ||
| − | |||
| − | <li class=" | + | |
| + | <li class="dropdown"><a class="dropdown-toggle" data-toggle="dropdown" href="#">Project </a> | ||
<ul class="dropdown-menu"> | <ul class="dropdown-menu"> | ||
<li><a href="https://2016.igem.org/Team:IISc_Bangalore/Description">Description</a></li> | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Description">Description</a></li> | ||
| − | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/ | + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Experiments">Experiments</a></li> |
| − | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/ | + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Results">Results</a></li> |
| − | + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Notebook">Notebook</a></li> | |
| − | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/ | + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Safety">Safety</a></li> |
</ul> | </ul> | ||
| − | </li> | + | </li> |
| + | |||
<li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Parts">Parts</a></li> | <li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Parts">Parts</a></li> | ||
| − | + | ||
<li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Collaborations">Collaborations</a></li> | <li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Collaborations">Collaborations</a></li> | ||
<li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Attributions">Attributions</a></li> | <li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/Attributions">Attributions</a></li> | ||
| − | <li class=" | + | |
| − | <li class="hvr-underline-from-left"><a | + | <li class="dropdown"><a class="dropdown-toggle" data-toggle="dropdown" href="#">Human Practices</a> |
| − | href="https://2016.igem.org/Team: | + | <ul class="dropdown-menu"> |
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Human_Practices">Human Practices</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/HP/Silver">HP Silver</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/HP/Gold">HP Gold</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li class="hvr-underline-from-left"><a href="https://2016.igem.org/Team:IISc_Bangalore/InterLab">InterLab</a></li> | ||
| + | <li class="dropdown"><a class="dropdown-toggle" data-toggle="dropdown" href="#">Special Prizes</a> | ||
| + | <ul class="dropdown-menu"> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Integrated_Practices">Integrated Human Practices</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Engagement">Education and Public Engagement</a></li> | ||
| + | <li class="active"><a href="https://2016.igem.org/Team:IISc_Bangalore/Model">Model</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Measurement">Measurement</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Entrepreneurship">Entrepreneurship</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:IISc_Bangalore/Design">Applied Design</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
</ul> | </ul> | ||
| Line 92: | Line 110: | ||
</div> | </div> | ||
</nav> | </nav> | ||
| + | |||
<div class="banner-main"> | <div class="banner-main"> | ||
| Line 128: | Line 147: | ||
<div id="welcome" class="welcome w3l"> | <div id="welcome" class="welcome w3l"> | ||
<div class="container"> | <div class="container"> | ||
| − | |||
| − | |||
<!-- accordian 1 --> | <!-- accordian 1 --> | ||
<div class="accordion" | <div class="accordion" | ||
| Line 139: | Line 156: | ||
<span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | <span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | ||
<h4>Overview</h4> | <h4>Overview</h4> | ||
| − | <p> | + | <p></p> |
</div> | </div> | ||
<div class="content"> | <div class="content"> | ||
| − | <p class="whitef">With the advent of rDNA technology in the late 1970s, medicine, agriculture and several other areas underwent a quantum leap | + | <p class="whitef">With the advent of rDNA technology in the late 1970s, medicine, agriculture and several other areas underwent a quantum leap</span><br> |
| − | + | <a href="#refer1" onclick="closeTab()"><span class="readm hvr-ripple-out">Read rest</span></a> | |
| + | </p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 150: | Line 168: | ||
<div class="heading blue"> | <div class="heading blue"> | ||
<span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | <span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | ||
| − | <h4> | + | <h4>Bioreactor design</h4> |
| − | + | ||
</div> | </div> | ||
<div class="content"> | <div class="content"> | ||
| − | + | As stated in the introduction, our project is an attempt to modify E coli to lower capital and running costs for rDNA bioreactor systems, by automating protein over-expression induction and separation of cell from growth media. | |
| − | To | + | To learn more about the existing biotech infrastructure in India, we decided to visit some biotech parks in India and explore the feasibility and applicability of our idea. |
| − | + | <br><a href="#refer2" onclick="closeTab()"><span class="readm hvr-ripple-out">Read rest</span></a> | |
| − | + | ||
| − | + | </p> | |
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
<div class="item col-md-3" data-anijs="if:mouseenter, on: .heading, do: pulse animated, to: $children target"> | <div class="item col-md-3" data-anijs="if:mouseenter, on: .heading, do: pulse animated, to: $children target"> | ||
<div class="heading blue"> | <div class="heading blue"> | ||
<span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | <span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | ||
| − | <h4>Protein production</h4> | + | <h4>Protein production module</h4> |
| − | + | ||
</div> | </div> | ||
| − | <div class="content | + | <div class="content">Induction synthesis of protein of interest is another important step. |
| − | + | ||
| − | + | <br><a href="#refer3" onclick="closeTab()"><span class="readm hvr-ripple-out">Read rest</span></a> | |
| − | + | ||
| − | </ | + | </p> |
</div> | </div> | ||
</div> | </div> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | <div class="item col-md-3" data-anijs="if:mouseenter, on: .heading, do: pulse animated, to: $children target"> | ||
<div class="heading blue"> | <div class="heading blue"> | ||
<span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | <span class="glyphicon glyphicon-chevron-down" aria-hidden="true" ></span> | ||
| − | <h4>Auto-aggregation | + | <h4>Auto-aggregation</h4> |
| − | + | ||
</div> | </div> | ||
<div class="content"> | <div class="content"> | ||
| − | <p class="whitef">To achieve cellular aggregation, we use the famous auto-transporter protein, Antigen43 (Ag43). <a href="# | + | <p class="whitef">To achieve cellular aggregation, we use the famous auto-transporter protein, Antigen43 (Ag43). |
| + | <br><a href="#refer4" onclick="closeTab()"><span class="readm hvr-ripple-out">Read more</span></a> | ||
| + | |||
</p> | </p> | ||
| − | |||
</div> | </div> | ||
| − | </div> | + | </div> |
| − | + | ||
| − | + | ||
</div> | </div> | ||
<!-- accordian 1 --> | <!-- accordian 1 --> | ||
| − | + | </div> | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<!--content-middle--> | <!--content-middle--> | ||
<div class="content-middle wthree"> | <div class="content-middle wthree"> | ||
| Line 257: | Line 232: | ||
<!--content-middle--> | <!--content-middle--> | ||
<div class="content-middle wthree"> | <div class="content-middle wthree"> | ||
| − | <div class="container" id=" | + | <div class="container" id="refer2"> |
<div class="mid-content wow fadeInRight animated" data-wow-delay=".5s"> | <div class="mid-content wow fadeInRight animated" data-wow-delay=".5s"> | ||
| − | <h3> | + | <h3>Bioreactor design</h3> |
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
<div class="refer col-md-6"> | <div class="refer col-md-6"> | ||
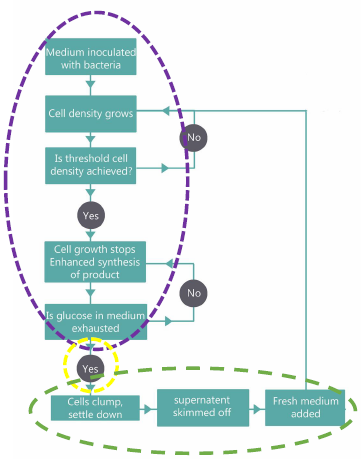
| − | < | + | <p class="whitef leftMove"></html>[[File:Flowchart iisc.jpg|right]]<html>We realized that manufacturing units use expensive and energy expensive machinery <a href="#refer" onclick="closeTab()">[3]</a>. Hence, we engineered our bacteria in such a way that it could do most of the work done by these monstrous machines so that we no longer have to buy them and pay their bills. |
| − | + | To increase the yield of the product, we came up with the idea of separating cell growth and target protein production so that when the cells have grown up to a high enough cell density, they shuttle all their energy towards the required protein production<a href="#refer" onclick="closeTab()"> [4]</a>. Based on the above ideas, we came up with the design of our bioreactor.<br> | |
| − | + | We realized that we would not be able to work on the whole idea this summer so, we focussed on two modules of our design: the Protein production module and the auto-aggregation module leaving the arresting of cell growth for future iGEM teams as a seed for their ideas. | |
| − | + | <!-- <br><span class="readm hvr-ripple-out">Read more</span> | |
| − | + | --> </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 286: | Line 252: | ||
<!--content-middle--> | <!--content-middle--> | ||
<div class="content-middle wthree"> | <div class="content-middle wthree"> | ||
| − | <div class="container" id=" | + | <div class="container" id="refer3"> |
<div class="mid-content wow fadeInRight animated" data-wow-delay=".5s"> | <div class="mid-content wow fadeInRight animated" data-wow-delay=".5s"> | ||
| − | <h3> | + | <h3>Protein production</h3> |
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
<div class="refer col-md-6"> | <div class="refer col-md-6"> | ||
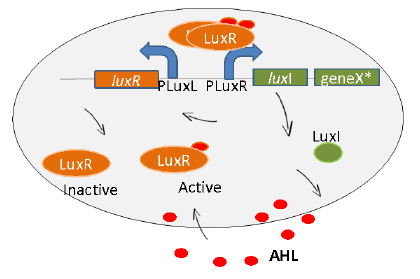
| − | < | + | <p class="whitef leftMove"></html>[[File:Lux iisc.jpg|right]]<html>Induction synthesis of protein of interest is another important step. Microorganisms in bioreactors are induced at the OD, at which the protein production is maximum. <a href="#refer" onclick="closeTab()">[5]</a> Because of this, factories install sophisticated machinery to keep track of the OD value of the culture so as to know when to induce the production. We wanted our bacteria to know when to start protein production and start it by itself. Hence, we used the lux quorum sensing system to sense this cell density. <a href="#refer" onclick="closeTab()">[6]</a> We have expressed the gene for our protein of interest under the luxI gene so that protein of interest is produced in significant quantities only when the quorum is achieved.(Figure 2) We also wish to tune the quorum sensing system by tweaking with the strength of the promoters involved in quorum sensing. |
| − | + | <!-- <br><span class="readm hvr-ripple-out">Read more</span> --> | |
| − | + | ||
| − | + | </p> </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 314: | Line 269: | ||
<!--content-middle--> | <!--content-middle--> | ||
<div class="content-middle wthree"> | <div class="content-middle wthree"> | ||
| − | <div class="container" id=" | + | <div class="container" id="refer4"> |
<div class="mid-content wow fadeInRight animated" data-wow-delay=".5s"> | <div class="mid-content wow fadeInRight animated" data-wow-delay=".5s"> | ||
| − | <h3> | + | <h3>Auto-aggregation</h3> |
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
<div class="refer col-md-6"> | <div class="refer col-md-6"> | ||
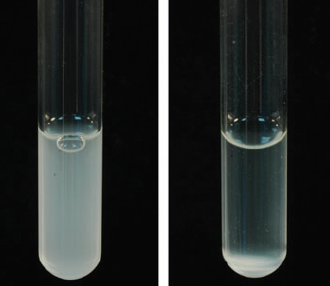
| − | < | + | <p class="whitef"></html>[[File:Tube iisc.jpg|right]]<html>To achieve cellular aggregation, we use the famous auto-transporter protein, Antigen43 (Ag43). <a href="#refer" onclick="closeTab()">[7]</a> It has shown to cause auto-aggregation of cells when overexpressed. (Figure 3) We plan to exploit this property of Ag43 to substitute centrifugation. We acknowledge previous iGEM teams who have worked on Ag43 (Hokkaido University, 2012; Aberdeen Scotland, 2014 and some more) as we got the BioBricks for Ag43 readily available. We intend to express Ag43 only when the cells have made the product of interest. Hence, we use the diauxic shift in the media to act as a switch for the expression of Ag43. |
| − | + | </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 01:06, 20 October 2016
Overview
With the advent of rDNA technology in the late 1970s, medicine, agriculture and several other areas underwent a quantum leap
Read rest
Bioreactor design
Read rest
Protein production module
Read rest
Auto-aggregation
To achieve cellular aggregation, we use the famous auto-transporter protein, Antigen43 (Ag43).
Read more
Overview
Bioreactor design
We realized that we would not be able to work on the whole idea this summer so, we focussed on two modules of our design: the Protein production module and the auto-aggregation module leaving the arresting of cell growth for future iGEM teams as a seed for their ideas.
Protein production
Auto-aggregation
References
- Ferrer-Miralles N, Domingo-Espin J, Corchero JL, Vazquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals, Microbial Cell Factories, 2009:8:17.
- Raisa Vermasvuori, PRODUCTION OF RECOMBINANT PROTEINS AND MONOCLONAL ANTIBODIES – TECHNO-ECONOMICAL EVALUATION OF THE PRODUCTION METHODS
- Felo M., Christensen B., Higgins J., Process cost and facility considerations in the selection of primary cell culture clarification technology, AIChE, 2013.
- Motoo Suzuki, Rohini Roy, Haiyan Zheng, Nancy Woychik, Masayori Inouye, Bacterial Bioreactors for High Yield Production of Recombinant Protein. The Journal of Biological Chemistry, 2006.
- Rosano G.L., Ceccarelli E. A., Recombinant protein expression in Escherichia coli: advances and challenges, Frontiers in Microbiology, 2014.
- Miller M.B., Bassler B.L., Quorum Sensing in Bacteria, Annual Review of Microbiology, 2001.
- Marjan W. van der Woude, Ian R. Henderson, Regulation and Function of Ag43 (Flu), Annu. Rev. Microbiol., 2008.
Overview
With the advent of rDNA technology in the late 1970s, medicine, agriculture and several other areas underwent a quantum leap and from that point, progress only hastened, from one only one recombinant pharmaceutical approved for human use (insulin) in 1982 to one hundred and fifty-one FDA approved protein based recombinant pharmaceuticals by 2009[1]. Despite being in high demand (due to the fact that most recombinant products produced on an industrial scale are therapies for chronic diseases like cancer and diabetes), recombinant products are expensive due to several factors like long and expensive development time, high failure rate (~80%) of the products developed, manufacturing costs requiring expensive technologies and processes (bioreactors, column chromatography, sterile conditions, etc) and the involvement of skilled labor on both the manufacturing and the healthcare provider’s side[2]. Treatments with these pharmaceuticals can cost from around 10,000 to 100,000 € per year for a single patient[2]. As scientists and engineers, it seems obvious that our contribution can be most easily and effectively be made at the level of manufacturing costs; to try to bring down the cost of these life-saving products.
Bioreactor design
We realized that manufacturing units use expensive and energy expensive machinery [3]. Hence, we engineered our bacteria in such a way that it could do most of the work done by these monstrous machines so that we no longer have to buy them and pay their bills.
To increase the yield of the product, we came up with the idea of separating cell growth and target protein production so that when the cells have grown up to a high enough cell density, they shuttle all their energy towards the required protein production [4]. Based on the above ideas, we came up with the design of our bioreactor.
We realized that we would not be able to work on the whole idea this summer so, we focussed on two modules of our design: the Protein production module and the auto-aggregation module leaving the arresting of cell growth for future iGEM teams as a seed for their ideas.
Protein production
Induction synthesis of protein of interest is another important step. Microorganisms in bioreactors are induced at the OD, at which the protein production is maximum. [5] Because of this, factories install sophisticated machinery to keep track of the OD value of the culture so as to know when to induce the production. We wanted our bacteria to know when to start protein production and start it by itself. Hence, we used the lux quorum sensing system to sense this cell density. [6] We have expressed the gene for our protein of interest under the luxI gene so that protein of interest is produced in significant quantities only when the quorum is achieved.(Figure 2) We also wish to tune the quorum sensing system by tweaking with the strength of the promoters involved in quorum sensing.
Auto-aggregation
To achieve cellular aggregation, we use the famous auto-transporter protein, Antigen43 (Ag43). [7] It has shown to cause auto-aggregation of cells when overexpressed. (Figure 3) We plan to exploit this property of Ag43 to substitute centrifugation. We acknowledge previous iGEM teams who have worked on Ag43 (Hokkaido University, 2012; Aberdeen Scotland, 2014 and some more) as we got the BioBricks for Ag43 readily available. We intend to express Ag43 only when the cells have made the product of interest. Hence, we use the diauxic shift in the media to act as a switch for the expression of Ag43.