| (63 intermediate revisions by 4 users not shown) | |||
| Line 20: | Line 20: | ||
</table> | </table> | ||
<span class="nomal2"> | <span class="nomal2"> | ||
| − | <br>We made a platform of technology for constructing covalently linked multi-enzyme-complex through disulfide bonds recruited by self-assembling peptide (SAP). By fusing SAP to | + | <br>We made a platform of technology for constructing covalently linked multi-enzyme-complex through disulfide bonds recruited by self-assembling peptide (SAP). By fusing SAP to ends of a protein, it will condense with other proteins’ SAP domains and form complexes. The SAP domains is pinched by short linkers (SLs) that have cysteine residues. When the SAPs gather and SLs get close, disulfide bonds are formed between two SLs. So, we will make an unbreakable complex. By using this method, we’ll be able to connect several enzymes and allow huge complexed proteins to be formed. It’ll improve the efficiency of a continuous reaction. |
</span> | </span> | ||
<br clear="all"> | <br clear="all"> | ||
| − | + | ||
| − | <table style="border-style: none"> | + | <table style="border-style: none; width:1px; margin-left:auto; margin-right:auto;"> |
<tr align="center"> | <tr align="center"> | ||
<td style="border-style: none;"> | <td style="border-style: none;"> | ||
| Line 35: | Line 35: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| − | + | ||
<br clear="all"> | <br clear="all"> | ||
</div> | </div> | ||
| Line 42: | Line 42: | ||
<br> | <br> | ||
| − | <br> | + | <br>Generally, linkers are used to connect proteins and make fusion proteins. We believe that our new method for connecting peptides with SAPs is superior to general methods using linkers for these reasons (Table. 1).</br> |
<br> | <br> | ||
<br> | <br> | ||
| + | |||
| + | </span> | ||
<div> | <div> | ||
| + | <center><span class="small">Table. 1. Comparison between linkers and SAPs</span></center> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<table class="merit"> | <table class="merit"> | ||
| − | |||
<tr> | <tr> | ||
<th width="50%">Linker Method</th> | <th width="50%">Linker Method</th> | ||
| Line 60: | Line 67: | ||
<tr> | <tr> | ||
| − | <td>Difficult to produce several huge | + | <td>Difficult to produce several huge complexes</td> |
<td>Possible to synthesize the proteins individually. Can also form a huge complex (Fig. 4)</td> | <td>Possible to synthesize the proteins individually. Can also form a huge complex (Fig. 4)</td> | ||
</tr> | </tr> | ||
| Line 90: | Line 97: | ||
<tr align="center" style="border-style: none"> | <tr align="center" style="border-style: none"> | ||
<td style="border-style: none;"><span class="small">Fig. 3. Using linkers<br>Expressions of gene A, B and C which code protein A, B and C are regulated by one promoter. If you connect some huge proteins, the expression efficiency may be decreased because the coding sequence is too long. </span></td> | <td style="border-style: none;"><span class="small">Fig. 3. Using linkers<br>Expressions of gene A, B and C which code protein A, B and C are regulated by one promoter. If you connect some huge proteins, the expression efficiency may be decreased because the coding sequence is too long. </span></td> | ||
| − | <td style="border-style: none;"><span class="small">Fig. 4. Using SAPs<br>You can produce protein A, B and C individually. After expression, they gather by SAPs and form | + | <td style="border-style: none;"><span class="small">Fig. 4. Using SAPs<br>You can produce protein A, B and C individually. After expression, they gather by SAPs and form disulfide bonds by SLs.</span></td> |
</tr> | </tr> | ||
</center> | </center> | ||
| Line 101: | Line 108: | ||
<br> | <br> | ||
<center> | <center> | ||
| − | <table style="border-style: none"> | + | <table style="border-style: none; width:1px; margin-left:auto; margin-right:auto;"> |
<tr align="center"> | <tr align="center"> | ||
<td style="border-style: none;"> | <td style="border-style: none;"> | ||
| Line 122: | Line 129: | ||
| − | + | ||
<table style="border-style: none"> | <table style="border-style: none"> | ||
<tr align="center" style="border-style: none"> | <tr align="center" style="border-style: none"> | ||
| − | <td style="border-style: none; "> | + | <td style="border-style: none; width:500px;"> |
| − | <img src="https://static.igem.org/mediawiki/2016/d/d8/T--HokkaidoU_Japan--multimerization_image5.png" alt="demerit" height="220px" width="auto"></td> | + | <img src="https://static.igem.org/mediawiki/2016/d/d8/T--HokkaidoU_Japan--multimerization_image5.png" alt="demerit" height="220px" width="auto"> |
| − | <td style="border-style: none; "> | + | </td> |
| − | <img src="https://static.igem.org/mediawiki/2016/c/cf/T--HokkaidoU_Japan--multimerization_image4.png" alt="resolution" height=" | + | <td style="border-style: none; width:500px;"> |
| − | </td> | + | <img src="https://static.igem.org/mediawiki/2016/c/cf/T--HokkaidoU_Japan--multimerization_image4.png" alt="resolution" height="300px" width="auto"> |
| + | </td> | ||
</tr> | </tr> | ||
<tr align="center" style="border-style: none"> | <tr align="center" style="border-style: none"> | ||
| − | <td style="border-style: none;"><span class="small">Fig. 6. Demerit of using SAP method<br>If some kinds of protein are expressed, | + | <td style="border-style: none; width:500px;"><span class="small">Fig. 6. Demerit of using SAP method<br>If some kinds of protein are expressed, there are so many combination. You may not be able to get the correct combination. </span></td> |
| − | <td style="border-style: none;"><span class="small">Fig. 7. Resolution for infinite combinations<br>When you use some kinds of SAP, | + | <td style="border-style: none; width:500px;"><span class="small">Fig. 7. Resolution for infinite combinations<br>When you use some kinds of SAP, incorrect connections will decrease.</span></td> |
</tr> | </tr> | ||
| − | |||
</table> | </table> | ||
| − | |||
<br clear="all"> | <br clear="all"> | ||
| + | </div> | ||
| + | |||
<span class="nomal2"> | <span class="nomal2"> | ||
| − | <br>Multimerization is very useful. As forming protein complex with different functions, this multimer let us create more functional units. When same kinds of | + | <br>Multimerization is very useful. As forming protein complex with different functions, this multimer let us create more functional units. When the same kinds of proteins are used, it’ll be a large block and its function is expected to be enhanced. |
<br> | <br> | ||
| − | <br>We tried to establish novel uses of SAP in this year. We tried | + | <br>We tried to establish novel uses of SAP in this year. We tried to multimerize using SAPs. |
</span> | </span> | ||
| − | |||
| + | |||
| + | |||
| + | <br> | ||
<br> | <br> | ||
<div id="Methods"><img src="https://static.igem.org/mediawiki/2016/2/2c/T--HokkaidoU_Japan--methods.png" | <div id="Methods"><img src="https://static.igem.org/mediawiki/2016/2/2c/T--HokkaidoU_Japan--methods.png" | ||
| Line 160: | Line 170: | ||
<br> | <br> | ||
| − | <br>We tried forming multimers using the self-assembling peptide (SAP), P<span class="sitatuki">11</span>-4 (QQRFEWEFEQQ) and RADA16-I (RADARADARADARADA). And to make firm bonds we designed short linker (GGCGG) called SL for short. We | + | <br>We tried forming multimers using the self-assembling peptide (SAP), P<span class="sitatuki">11</span>-4 (QQRFEWEFEQQ) and RADA16-I (RADARADARADARADA). And to make firm bonds we designed a short linker (GGCGG) called SL for short. We |
| − | + | connected SL and SAP to both ends of a protein. In this experiment, we used GFP as a fusion partner (Fig. 8). | |
| − | + | </sapn> | |
<table style="border-style: none"> | <table style="border-style: none"> | ||
| Line 188: | Line 198: | ||
</table> | </table> | ||
| − | <br>GFP’s molecular | + | <br>GFP’s molecular weight is 26891 Da. When GFP is fused with P<span class="sitatuki">11</span>-4, the molecular weight of the fusion protein is estimated to be 31709 Da. Fused with RADA16-I, it’s 31943 Da. When they form a multimer, the molecular weight will be more |
| − | 31709 Da. | + | than 60 kDa. Consequently, we used the filter which filters out the proteins with weight of more than 50 KDa. |
| − | than 60 kDa. Consequently, we used the filter which filters out the proteins with | + | |
<br>For the evaluation, we ordered IDT the designed constructions and put them on the vectors. Then, | <br>For the evaluation, we ordered IDT the designed constructions and put them on the vectors. Then, | ||
| − | we introduced them | + | we introduced them into <span style="font-style: italic">E.coli</span> and the protein expression was induced with IPTG. Causing bacteriolysis with freeze-thaw, we acquired the supernatant containing the proteins by centrifugal |
separation. Purifying the protein with Ni-affinity chromatography, we filtrated the solution | separation. Purifying the protein with Ni-affinity chromatography, we filtrated the solution | ||
| − | to separate the proteins with | + | to separate the proteins with weight of less than 50KDa. We irradiated 480 nm light to filtrate and observed |
whether 580 nm wave-length light was emitted.<br> | whether 580 nm wave-length light was emitted.<br> | ||
</div> | </div> | ||
| Line 203: | Line 212: | ||
<br> | <br> | ||
| − | + | <div id="Results"><img src="https://static.igem.org/mediawiki/2016/d/de/T--HokkaidoU_Japan--results.png" | |
width="250px" height="90px" alt="results"></div></p> | width="250px" height="90px" alt="results"></div></p> | ||
| Line 209: | Line 218: | ||
| − | <br>We | + | <br>We inserted SAP-GFP-SAP coding reasion (Fig. 8) into pET15b vector and the fusion protein was expressed (Fig. 10). As a negative control we made a construct containing GFP alone (Fig. 11). GFPs with SAPs and SLs was expected to form multiple complexes (Fig. 12). |
</span> | </span> | ||
| + | |||
| + | |||
| + | |||
| + | <center> | ||
<table style="border-style: none"> | <table style="border-style: none"> | ||
| − | <tr align="center"> | + | <tr align="center" style="border-style: none"> |
| − | <td style="border-style: none;"> | + | <td style="border-style: none; width:500px;"> |
| − | + | <img src="https://static.igem.org/mediawiki/2016/d/d7/T--HokkaidoU_Japan--Multimerization_Construct.png" alt="construct" height="180px" width="auto"> | |
| − | <td style="border-style:none; | + | </td> |
| − | + | <td style="border-style: none; width:500px;"> | |
| − | + | <img src="https://static.igem.org/mediawiki/2016/e/e8/T--HokkaidoU_Japan--Multimerization_ConstructNC.png" alt="Negative control" height="180px" width="auto"> | |
| − | <td style="border-style: none | + | </td> |
| + | </tr> | ||
| + | |||
| + | |||
| + | <tr align="center" style="border-style: none"> | ||
| + | <td style="border-style: none; width:500px;"><span class="small">Fig. 10. A construct of multimerization using SAP<br>This is the construct for making multiple complex. We used RADA16-I and P<span class="sitatuki">11</span>-4 as SAP. <br>C is a cysteine residues in short linker.</span></td> | ||
| + | <td style="border-style: none; width:500px;"><span class="small">Fig. 11. A construct of a negative control<br>We made a negative control which had only GFP to test the effect of SAPs.</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | </center> | ||
| + | |||
<br clear="all"> | <br clear="all"> | ||
| − | <table style="border-style: none"> | + | |
| + | <div> | ||
| + | <table style="border-style: none;float:right"> | ||
| + | |||
<tr align="center"> | <tr align="center"> | ||
<td style="border-style: none;"> | <td style="border-style: none;"> | ||
<tr> | <tr> | ||
| − | <td style="border-style:none; float:center"><img src="https://static.igem.org/mediawiki/2016/ | + | <td style="border-style:none; float:center"><img src="https://static.igem.org/mediawiki/2016/2/28/T--HokkaidoU_Japan--image21.png" alt="image" height="500px" width="auto"></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center"><span class="small">Fig. | + | <td style="border-style: none"; align="center"><span class="small">Fig. 12. Expected forming multiple complex</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | <span class="nomal2"> | ||
| + | <br>We planed to express GFPs fused with SAPs and form disulfide bonds by glutathione-S-transferase (GST) (Fig. 12). If GFPs form multiple complexes, we would reveal that SAPs could recruit proteins. But we couldn't get the colony that had SAP-GFP-SAP on pET15b vector. | ||
| + | <br>So, we changed our strategy to making other constructs, that are, <a href="http://parts.igem.org/Part:BBa_K2015012">BBa_K2015012</a>-<a href="http://parts.igem.org/Part:BBa_K2015008">BBa_K2015008</a> on pSB1C3 and <a href="http://parts.igem.org/Part:BBa_K2015012">BBa_K2015012</a>-<a href="http://parts.igem.org/Part:BBa_K2015009">BBa_K2015009</a> on pSB1C3. Our biobrick <a href="http://parts.igem.org/Part:BBa_K2015012">BBa_K2015012</a> was designed to code LacI and have PLac. Both <a href="http://parts.igem.org/Part:BBa_K2015008">BBa_K2015008</a> and <a href="http://parts.igem.org/Part:BBa_K2015009">BBa_K2015009</a> code SAP-GFP-SAP, but former has RADA16-I parts and latter has P<span class="sitatuki">11</span>-4 parts. | ||
| + | <br>We inserted them into pSB1C3 vector and verified whether our device went well. But the fluorescence of GFPs was not detected any condition (Fig. 13 and 14). | ||
| + | </span> | ||
| + | |||
| + | </div> | ||
<br clear="all"> | <br clear="all"> | ||
| − | + | ||
| − | <table style="border-style: none"> | + | |
| + | |||
| + | <table style="border-style: none; width:1px; margin-left:auto; margin-right:auto"> | ||
<tr align="center"> | <tr align="center"> | ||
<td style="border-style: none;"> | <td style="border-style: none;"> | ||
<tr> | <tr> | ||
| − | <td style="border-style:none; float:center"><img src="https://static.igem.org/mediawiki/2016/ | + | <td style="border-style:none; float:center"><img src="https://static.igem.org/mediawiki/2016/d/da/T--HokkaidoU_Japan--multimerization_BBa_K2015012-BBa_K2015008.png" alt="result1" height="300px" width="auto"></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center"><span class="small">Fig. | + | <td style="border-style: none"; align="center"><span class="small">Fig. 13. BBa_K2015012-BBa_K2015008 induced by IPTG<br>BBa_K2015012-BBa_K2015008 on pSB1C3 vector were induced by IPTG and incubated at 37°C. (-): not induced, (+): 0.5 mM IPTG and (++): 1.0 mM IPTG</span></td> |
</tr> | </tr> | ||
| + | </tr> | ||
</table> | </table> | ||
<br clear="all"> | <br clear="all"> | ||
| + | |||
| + | |||
| + | <center> | ||
| + | <table style="border-style: none"> | ||
| + | <tr align="center" style="border-style: none"> | ||
| + | <td style="border-style: none; "> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/b/be/T--HokkaidoU_Japan--multimerization_BBa_K2015012-BBa_K2015009_25.png" alt="result2" height="300px" width="auto"> | ||
| + | </td> | ||
| + | <td style="border-style: none; "> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/f/fa/T--HokkaidoU_Japan--multimerization_BBa_K2015012-BBa_K2015009.png" alt="result3" height="300px" width="auto"> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | <span class="small">Fig. 14. BBa_K2015012-BBa_K2015009 induced by IPTG<br>BBa_K2015012-BBa_K2015009 on pSB1C3 vector were induced by IPTG and incubated at 25°C for 24 h (left) <br>and at 37°C for 16 h (right). (-): not induced, (+): 0.5 mM IPTG and (++): 1.0 mM IPTG</span> | ||
| + | </center> | ||
| + | <br clear="all"> | ||
| + | |||
| + | |||
| Line 262: | Line 315: | ||
<span class="nomal2"> | <span class="nomal2"> | ||
| − | <br> | + | |
| + | <br><a href="http://parts.igem.org/Part:BBa_K2015012">BBa_K2015012</a> codes constitutive promoter, RBS, LacI, dT, PLac and RBS. When RFP was inserted to it, we could see fluorescence (<a href="https://2016.igem.org/Team:HokkaidoU_Japan/Proof">Proof</a>). So, this units would be well. We thought GFPs were expressed but their functions were lost. There is a possibility of forming inclusion bodies because of SAPs. We need to examine whether GFPs are produced using SDS-PAGE. | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <br>Our constructs have two <span style="font-style: italic">Bam</span>HI sites at upstream and downstream of GFP. Therefore, anyone can cut out the GFP and insert any protein of interest which has <span style="font-style: italic">Bam</span>HI sites at both ends. our construct permits anyone to make multi-complex-enzyme(Fig. 15). | ||
</span> | </span> | ||
| − | <table style="border-style: none"> | + | <table style="border-style: none; width:1px; margin-left:auto; margin-right:auto"> |
<tr align="center"> | <tr align="center"> | ||
<td style="border-style: none;"> | <td style="border-style: none;"> | ||
| Line 274: | Line 332: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="border-style: none"; align="center"><span class="small">Fig. | + | <td style="border-style: none"; align="center"><span class="small">Fig. 15. A construct for making novel artificial multi-enzyme-complex<br>We designed this construct to have a cloning site. If you design the protein which has <span style="font-style: italic">Bam</span>HI sites, you can make a multimerized protein.</span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 287: | Line 345: | ||
<span class="nomal2"> | <span class="nomal2"> | ||
| − | <br>[1] Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242 | + | <br>[1] Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242-251. |
<br>[2] Castellana M1, Wilson MZ2, Xu Y3, Joshi P2, Cristea IM2, Rabinowitz JD4, Gitai Z2, Wingreen NS3. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014 Oct;32(10):1011-8. | <br>[2] Castellana M1, Wilson MZ2, Xu Y3, Joshi P2, Cristea IM2, Rabinowitz JD4, Gitai Z2, Wingreen NS3. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014 Oct;32(10):1011-8. | ||
</span> | </span> | ||
| − | + | ||
</div> <!-- 2016contents 閉じる --> | </div> <!-- 2016contents 閉じる --> | ||
<!--begin footer--> | <!--begin footer--> | ||
Latest revision as of 03:04, 20 October 2016
Team:HokkaidoU Japan
\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\\

We made a platform of technology for constructing covalently linked multi-enzyme-complex through disulfide bonds recruited by self-assembling peptide (SAP). By fusing SAP to ends of a protein, it will condense with other proteins’ SAP domains and form complexes. The SAP domains is pinched by short linkers (SLs) that have cysteine residues. When the SAPs gather and SLs get close, disulfide bonds are formed between two SLs. So, we will make an unbreakable complex. By using this method, we’ll be able to connect several enzymes and allow huge complexed proteins to be formed. It’ll improve the efficiency of a continuous reaction.
Generally, linkers are used to connect proteins and make fusion proteins. We believe that our new method for connecting peptides with SAPs is superior to general methods using linkers for these reasons (Table. 1).
Table. 1. Comparison between linkers and SAPs
We thought the SAP method was best one but it also had disadvantages. Since the number of the possible combination of several different proteins is infinite, there is no guarantee that we can always obtain the expected combination.
One solution to the problem is limiting the number of combination by using different SAP. That can reduce probability of incorrect connection a little.
Multimerization is very useful. As forming protein complex with different functions, this multimer let us create more functional units. When the same kinds of proteins are used, it’ll be a large block and its function is expected to be enhanced.
We tried to establish novel uses of SAP in this year. We tried to multimerize using SAPs.

We tried forming multimers using the self-assembling peptide (SAP), P11-4 (QQRFEWEFEQQ) and RADA16-I (RADARADARADARADA). And to make firm bonds we designed a short linker (GGCGG) called SL for short. We connected SL and SAP to both ends of a protein. In this experiment, we used GFP as a fusion partner (Fig. 8).
GFP’s molecular weight is 26891 Da. When GFP is fused with P11-4, the molecular weight of the fusion protein is estimated to be 31709 Da. Fused with RADA16-I, it’s 31943 Da. When they form a multimer, the molecular weight will be more than 60 kDa. Consequently, we used the filter which filters out the proteins with weight of more than 50 KDa.
For the evaluation, we ordered IDT the designed constructions and put them on the vectors. Then, we introduced them into E.coli and the protein expression was induced with IPTG. Causing bacteriolysis with freeze-thaw, we acquired the supernatant containing the proteins by centrifugal separation. Purifying the protein with Ni-affinity chromatography, we filtrated the solution to separate the proteins with weight of less than 50KDa. We irradiated 480 nm light to filtrate and observed whether 580 nm wave-length light was emitted.

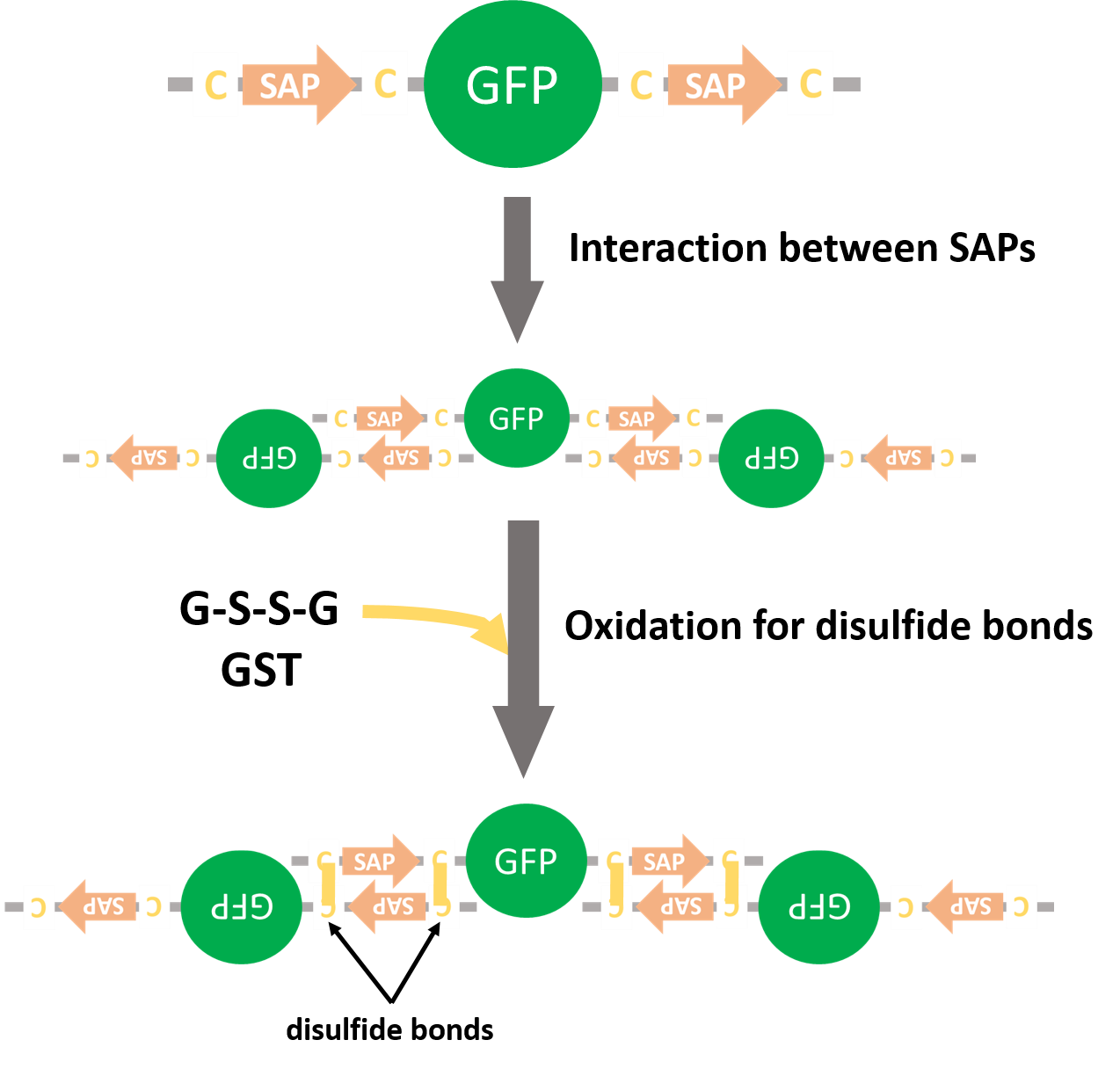
We inserted SAP-GFP-SAP coding reasion (Fig. 8) into pET15b vector and the fusion protein was expressed (Fig. 10). As a negative control we made a construct containing GFP alone (Fig. 11). GFPs with SAPs and SLs was expected to form multiple complexes (Fig. 12).
We planed to express GFPs fused with SAPs and form disulfide bonds by glutathione-S-transferase (GST) (Fig. 12). If GFPs form multiple complexes, we would reveal that SAPs could recruit proteins. But we couldn't get the colony that had SAP-GFP-SAP on pET15b vector.
So, we changed our strategy to making other constructs, that are, BBa_K2015012-BBa_K2015008 on pSB1C3 and BBa_K2015012-BBa_K2015009 on pSB1C3. Our biobrick BBa_K2015012 was designed to code LacI and have PLac. Both BBa_K2015008 and BBa_K2015009 code SAP-GFP-SAP, but former has RADA16-I parts and latter has P11-4 parts.
We inserted them into pSB1C3 vector and verified whether our device went well. But the fluorescence of GFPs was not detected any condition (Fig. 13 and 14).
Fig. 14. BBa_K2015012-BBa_K2015009 induced by IPTG
BBa_K2015012-BBa_K2015009 on pSB1C3 vector were induced by IPTG and incubated at 25°C for 24 h (left)
and at 37°C for 16 h (right). (-): not induced, (+): 0.5 mM IPTG and (++): 1.0 mM IPTG

BBa_K2015012 codes constitutive promoter, RBS, LacI, dT, PLac and RBS. When RFP was inserted to it, we could see fluorescence (Proof). So, this units would be well. We thought GFPs were expressed but their functions were lost. There is a possibility of forming inclusion bodies because of SAPs. We need to examine whether GFPs are produced using SDS-PAGE.
Our constructs have two BamHI sites at upstream and downstream of GFP. Therefore, anyone can cut out the GFP and insert any protein of interest which has BamHI sites at both ends. our construct permits anyone to make multi-complex-enzyme(Fig. 15).
[1] Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242-251.
[2] Castellana M1, Wilson MZ2, Xu Y3, Joshi P2, Cristea IM2, Rabinowitz JD4, Gitai Z2, Wingreen NS3. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014 Oct;32(10):1011-8.

 |
| Fig. 1. The enzyme reaction by multiple complex To connect different enzymes will make continuous reaction efficiently. |
We made a platform of technology for constructing covalently linked multi-enzyme-complex through disulfide bonds recruited by self-assembling peptide (SAP). By fusing SAP to ends of a protein, it will condense with other proteins’ SAP domains and form complexes. The SAP domains is pinched by short linkers (SLs) that have cysteine residues. When the SAPs gather and SLs get close, disulfide bonds are formed between two SLs. So, we will make an unbreakable complex. By using this method, we’ll be able to connect several enzymes and allow huge complexed proteins to be formed. It’ll improve the efficiency of a continuous reaction.
 |
| Fig. 2. Huge complex using SAP To connect same enzymes like fluorescent proteins will amplify their effects. |
Generally, linkers are used to connect proteins and make fusion proteins. We believe that our new method for connecting peptides with SAPs is superior to general methods using linkers for these reasons (Table. 1).
| Linker Method | SAP Method |
|---|---|
| Regulated by one promoter (Fig. 3) | Each protein can be produced individually (Fig. 4) |
| Difficult to produce several huge complexes | Possible to synthesize the proteins individually. Can also form a huge complex (Fig. 4) |
| The possibility of deformation of the 3D-structure (Fig. 5) | Low possibility of deformation since they only connect with proteins which can condense |

|

|
| Fig. 3. Using linkers Expressions of gene A, B and C which code protein A, B and C are regulated by one promoter. If you connect some huge proteins, the expression efficiency may be decreased because the coding sequence is too long. |
Fig. 4. Using SAPs You can produce protein A, B and C individually. After expression, they gather by SAPs and form disulfide bonds by SLs. |
 |
| Fig. 5. Demerit of using linkers In linker method, you need to consider the linker length to avoid the steric hindrance. |
We thought the SAP method was best one but it also had disadvantages. Since the number of the possible combination of several different proteins is infinite, there is no guarantee that we can always obtain the expected combination.
One solution to the problem is limiting the number of combination by using different SAP. That can reduce probability of incorrect connection a little.

|

|
| Fig. 6. Demerit of using SAP method If some kinds of protein are expressed, there are so many combination. You may not be able to get the correct combination. |
Fig. 7. Resolution for infinite combinations When you use some kinds of SAP, incorrect connections will decrease. |
Multimerization is very useful. As forming protein complex with different functions, this multimer let us create more functional units. When the same kinds of proteins are used, it’ll be a large block and its function is expected to be enhanced.
We tried to establish novel uses of SAP in this year. We tried to multimerize using SAPs.

We tried forming multimers using the self-assembling peptide (SAP), P11-4 (QQRFEWEFEQQ) and RADA16-I (RADARADARADARADA). And to make firm bonds we designed a short linker (GGCGG) called SL for short. We connected SL and SAP to both ends of a protein. In this experiment, we used GFP as a fusion partner (Fig. 8).
 |
| Fig. 8. Design of the coding sequence |
Assay
 |
| Fig. 9. Method for verifying whether proteins form multiple complex |
GFP’s molecular weight is 26891 Da. When GFP is fused with P11-4, the molecular weight of the fusion protein is estimated to be 31709 Da. Fused with RADA16-I, it’s 31943 Da. When they form a multimer, the molecular weight will be more than 60 kDa. Consequently, we used the filter which filters out the proteins with weight of more than 50 KDa.
For the evaluation, we ordered IDT the designed constructions and put them on the vectors. Then, we introduced them into E.coli and the protein expression was induced with IPTG. Causing bacteriolysis with freeze-thaw, we acquired the supernatant containing the proteins by centrifugal separation. Purifying the protein with Ni-affinity chromatography, we filtrated the solution to separate the proteins with weight of less than 50KDa. We irradiated 480 nm light to filtrate and observed whether 580 nm wave-length light was emitted.

We inserted SAP-GFP-SAP coding reasion (Fig. 8) into pET15b vector and the fusion protein was expressed (Fig. 10). As a negative control we made a construct containing GFP alone (Fig. 11). GFPs with SAPs and SLs was expected to form multiple complexes (Fig. 12).

|

|
| Fig. 10. A construct of multimerization using SAP This is the construct for making multiple complex. We used RADA16-I and P11-4 as SAP. C is a cysteine residues in short linker. |
Fig. 11. A construct of a negative control We made a negative control which had only GFP to test the effect of SAPs. |
 |
| Fig. 12. Expected forming multiple complex |
We planed to express GFPs fused with SAPs and form disulfide bonds by glutathione-S-transferase (GST) (Fig. 12). If GFPs form multiple complexes, we would reveal that SAPs could recruit proteins. But we couldn't get the colony that had SAP-GFP-SAP on pET15b vector.
So, we changed our strategy to making other constructs, that are, BBa_K2015012-BBa_K2015008 on pSB1C3 and BBa_K2015012-BBa_K2015009 on pSB1C3. Our biobrick BBa_K2015012 was designed to code LacI and have PLac. Both BBa_K2015008 and BBa_K2015009 code SAP-GFP-SAP, but former has RADA16-I parts and latter has P11-4 parts.
We inserted them into pSB1C3 vector and verified whether our device went well. But the fluorescence of GFPs was not detected any condition (Fig. 13 and 14).
 |
| Fig. 13. BBa_K2015012-BBa_K2015008 induced by IPTG BBa_K2015012-BBa_K2015008 on pSB1C3 vector were induced by IPTG and incubated at 37°C. (-): not induced, (+): 0.5 mM IPTG and (++): 1.0 mM IPTG |

|

|
BBa_K2015012-BBa_K2015009 on pSB1C3 vector were induced by IPTG and incubated at 25°C for 24 h (left)
and at 37°C for 16 h (right). (-): not induced, (+): 0.5 mM IPTG and (++): 1.0 mM IPTG

BBa_K2015012 codes constitutive promoter, RBS, LacI, dT, PLac and RBS. When RFP was inserted to it, we could see fluorescence (Proof). So, this units would be well. We thought GFPs were expressed but their functions were lost. There is a possibility of forming inclusion bodies because of SAPs. We need to examine whether GFPs are produced using SDS-PAGE.
Our constructs have two BamHI sites at upstream and downstream of GFP. Therefore, anyone can cut out the GFP and insert any protein of interest which has BamHI sites at both ends. our construct permits anyone to make multi-complex-enzyme(Fig. 15).
 |
| Fig. 15. A construct for making novel artificial multi-enzyme-complex We designed this construct to have a cloning site. If you design the protein which has BamHI sites, you can make a multimerized protein. |
[1] Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242-251.
[2] Castellana M1, Wilson MZ2, Xu Y3, Joshi P2, Cristea IM2, Rabinowitz JD4, Gitai Z2, Wingreen NS3. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014 Oct;32(10):1011-8.

 "
"





