| Line 3: | Line 3: | ||
'''In order to guarantee the efficiency of our own secreted enzymes and find optimum environment in our project set up for the own enzymes and bought ones, a functional enzyme assay is necessary.''' | '''In order to guarantee the efficiency of our own secreted enzymes and find optimum environment in our project set up for the own enzymes and bought ones, a functional enzyme assay is necessary.''' | ||
| − | |||
== Introduction == | == Introduction == | ||
| Line 24: | Line 23: | ||
glycosidic linkages into monosaccahrides such as beta-glucose. Xylanase breaks down | glycosidic linkages into monosaccahrides such as beta-glucose. Xylanase breaks down | ||
hemicellulose by degrading polysaccahride beta-1,4-xylan (Beechwood) into xylose. | hemicellulose by degrading polysaccahride beta-1,4-xylan (Beechwood) into xylose. | ||
| − | |||
== Experimental Set Up an Validation == | == Experimental Set Up an Validation == | ||
| Line 37: | Line 35: | ||
DNS is aromatic compound that reacts with reducing sugars to form 3-amino-5-nitrosalicylic acid- | DNS is aromatic compound that reacts with reducing sugars to form 3-amino-5-nitrosalicylic acid- | ||
| + | {{UBonn_HBRS/image|url=//2016.igem.org/wiki/images/6/6a/T--UBonn_HBRS--Enzyme_Assays_Kyoto.jpg|alt=Kyoto digestion DNS assay|class=100|caption-class=with-caption|caption=<i>iGEM</i>}} | ||
| + | |||
| + | This absorbs light at 540 nm creating a possibility to quantify the product present in the sample. | ||
| + | Additionally, absorbance at 750 nm was measured in order to subtract the background absorbance | ||
| + | for normalization. <br /> | ||
| + | Additional water served purpose for having room to change different part of the recipe while still | ||
| + | keeping constant volume. <br /> | ||
| + | Analysis of the data was done with Open Office Calc. | ||
{{UBonn_HBRS/footer}} | {{UBonn_HBRS/footer}} | ||
Revision as of 04:30, 19 October 2016
Enzyme Assays

In order to guarantee the efficiency of our own secreted enzymes and find optimum environment in our project set up for the own enzymes and bought ones, a functional enzyme assay is necessary.
Introduction
The aim of the enzyme assays, is to determine the amount of enzymes present in the sample under certain conditions, making activity comparable between the samples and experiments. The activity is described by the product formed over the given time or how much substrate has been used up.
Different parameters affecting the activity of an enzymes are substrates and their concentration, pH, ionic strength, buffer type and temperature.
In our project, method of stopped colorimetric assays was used. This means that the assay was stopped after a fixed time with the use of temperature and the amount of product formed was measured. Colorimetric assay makes it possible to to quantify the product of interest with the use of spectrophotometer, by reacting it with another reagent. The measured absorbance is directly proportional to the product in the sample. In this experiment, 3,5-Dinitrosalicylic acid was used.
The enzymes tested were endoglucanase (endo-1,4-β-D-glucanase, Acidothermus cellulolyticus, Sigma-Aldrich) and xylanase (endo-1,4-β-Xylanase M4, Aspergillus niger, Megazyme). First enzyme belongs to cellalases and breaks down cellulose molecule by hydrolysis of the 1,4-beta-D- glycosidic linkages into monosaccahrides such as beta-glucose. Xylanase breaks down hemicellulose by degrading polysaccahride beta-1,4-xylan (Beechwood) into xylose.
Experimental Set Up an Validation
The tests done always follow similar recipe (see protocol) that containing sugar or enzyme solution, citrate buffer, water, substrate and DNS. Buffers are needed to adjust and stabilize the pH for enzyme assays. The concentration that works for our chosen enzymes is 50mM. Substrate used for endoglucanase was Carboxymethyl cellulose (CMC) which is a cellulose derivative that the enzyme is able to cleave. Initially for the standard calibrations 1:3 CMC was used but in order to increase the availability of the substrate, the assays with enzymes had CMC 3:7. Substrate for xylanase was 1% xylan, also known as beechwood. DNS is aromatic compound that reacts with reducing sugars to form 3-amino-5-nitrosalicylic acid-
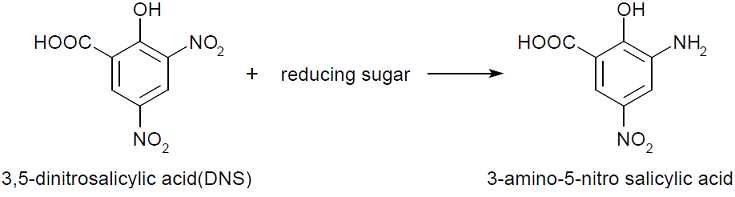
This absorbs light at 540 nm creating a possibility to quantify the product present in the sample.
Additionally, absorbance at 750 nm was measured in order to subtract the background absorbance
for normalization.
Additional water served purpose for having room to change different part of the recipe while still
keeping constant volume.
Analysis of the data was done with Open Office Calc.


