L.lactis Based Oral Vaccination: Presentation of expressed antigen to Antigen Presenting Cells

Introduction of reporter system:
The aim of this experiment is to produce a tool for in vitro experiments that will reveal if proteins are being delivered to particular cells. One of our initial investigations was whether L.lactis could produce proteins that are functional in the cell. Standard fluorescent proteins are able to fluoresce regardless if they’re expressed by bacteria or mammalian cells. Therefore, we would be unable to clarify if the protein is functional inside mammalian cells as phagosome internalised bacteria would also fluoresce once they express this protein. Ideally, we would be interested in using a protein in our reporter system that doesn’t produce a readout when it has been expressed by bacteria but once in a mammalian environment it could. This may be due to post-translational modifications such as glycosylation or the presence of co-factors in the cytosol of mammalian cells. For our reporter system, we have chosen a protein that can produce a readout because of the latter option ie requires a cofactor. The IRFP protein was developed as a reporter tool in our project.
Introduction on IRFP protein
iRFP is an infrared fluorescent protein that requires an excitation wavelength of approx. peak 660 nm and an emission wavelength of approx. 710 nm. The iRFP protein was used in the experiment which is slightly red-shifted when compared with iRFP 1.4 (Filonov et al.).

Figure 1-iRFP excitation and emission spectrum
iRFP requires the presence of a co-factor billiverdin to fluoresce. Billiverdin is formed through the breakdown of haeme and is present in the serum of blood and in the cytosol of mammalian cells. Billiverdin is normally converted to bilirubin via biliverdin reductase and lipophilic reactive species catalyse the reverse reaction.

Figure 2-Interconversion of Bilirubin to Biliverdin and Heme degradation (Sedlak & Snyder)
Biliverdin acts as the phytochrome for the iRFP protein and is therefore required for fluorescence. Utilising this information, it would be expected that bacterial expression of iRFP would not be fluorescent since there is no biliverdin present whereas if the bacteria had a source of a biliverdin it would. Biliverdin is not present in normal bacterial media and so L.lactis in GM17 would not be fluorescent . It is however present in cell culture media as it is in foetal bovine serum or FBS. As mentioned previously biliverdin is also present in the cytosol of mammalian cells. Therefore it is possible to use iRFP as a reporter system for showing whether L.lactis could produce protein in the cytosol of a macrophage cell line. The macrophage would phagocytose L.lactis and L.lactis would now be present in a phagosome. The listerolysin protein allows for phagosomal escape and for L.lactis to enter the cytosol of the cell.
RAW 264.7, a mouse macrophage cell line, were used in this experiment. Co-incubation of the L.lactis strain with the macrophages led to phagocytosis of the bacteria. An odyssey scanner can be used to image the results. Our procedure consisted of seeding the macrophages at a density of 1 x 105 cells per well in a 12 well plate. The total amount of culture medium in each well was 1 ml. The RAW 264.7 cells were allowed to grow for four hours before bacteria was added to the wells. Simultaneously, L.lactis was subcultured from a fresh overnight and grown to an OD of approx 1. Three different multiplicity of infections were used in the experiments (100:1, 500:1, 1000:1 in the initial experiments and 100:1, 300:1, 1000:1 in the later experiments). An OD to CFU curve was created for this strain of L.lactis in order to convert the OD 600 reading into the number of colony forming units. Following addition, the RAW 264.7 cells and L.lactis were left at 37 degrees Celsius 5 % CO2 for 4 hours . After this co-incubation penicillin/ streptomycin was added to each of the wells to kill the extracellular and intracellular bacteria and the plate was incubated for a further 1-2 hours. A PBS wash was carried out and the plate was imaged in the Odyssey scanner.
IRFP construct
The plasmid we used in our experiments had to be capable of replicating in L.lactis. We also needed to ensure that there was suitable antibiotic resistance. One of the plasmids that was previously used with L.lactis by groups in our university is PNZ44. This plasmid contains the P44 promoter which is a strong promoter in L.lactis. It also has chloramphenicol resistance gene, RepA and RepC replication initiation genes. The annotated XbaI restriction site was used to digest the plasmid before cloning in our constructs of interest. We used Gibson cloning to ligate in the IRFP construct into this site which is downstream of the P44 promoter.

Figure 3-PNZ44 plasmid on Benchling
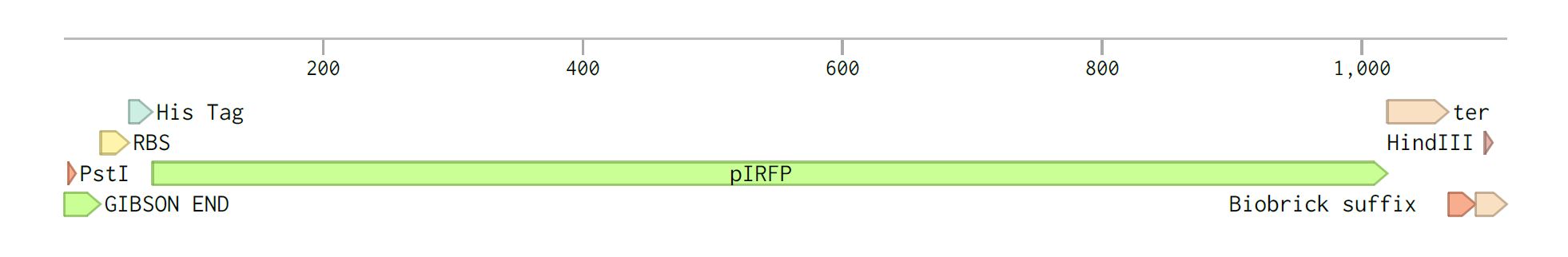
Figure 4-iRFP construct on Benchling

Figure 5-pNZ44-iRFP on Benchling
iRFP validation experiment- initial trials

iRFP validation - altered experiment

Results

Figure 6-Well 1: 100:1 MOI. Well 2 500:1 MOI. Well 3 1000:1 MOI. Well 4: RAW 264.7 cells only without bacteria. Analysis was carried out after the initial 1-2 hours following antibiotic addition.

Image analysis was carried out after 1-2 incubation following antibiotic addition. In this early trial we were interested in analysing how long before the protein produced a fluorescent signal and determining how long this signal will last before the protein is degraded. Since the addition of antibiotic will kill any bacteria and IRFP can only be expressed by bacteria, it is hypothesised that the protein signal will be reduced over time because of possible proteasomal degradation.
A further analysis was taken 12 hours after the addition of the antibiotic.
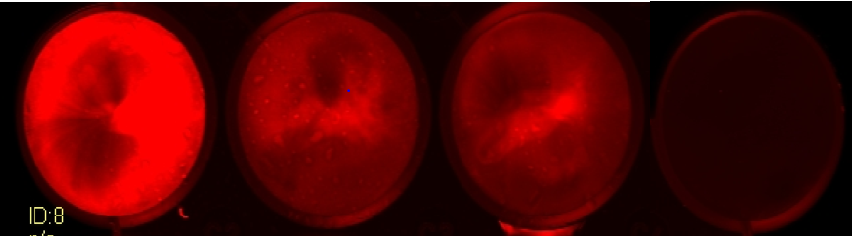
Figure 7-Well 1-100:1 MOI. Well 2-500:1 MOI. Well 3- 1000:1 MOI. Well 4 RAW 264.7 only.

Following another 12 hours, the 100:1 MOI has the highest MOI. This suggests that the higher number of L.lactis might cause irreversible cell stress for the macrophages during the co-incubation period. L.lactis produce lactic acid in metabolism which will lower the pH of the medium and this may trigger macrophage cell stress. An alternative hypothesis is that the protein is being degraded by normal degradation pathways in the cytosol of the macrophage.
However, for the above experiment, the RAW 264.7 cells were grown in DMEM & FBS. As mentioned previously biliverdin can be obtained from FBS through bilirubin which is present at a range of 4-11 % in the serum

Figure 7: SEQ Figure \* ARABIC 8-FBS composition
Due to the presence of bilirubin in FBS, our subsequent experiments focused on analysing whether RAW 264.7 cells grown in media without FBS for the duration of the experiment would produce fluorescence. The cells were grown in DMEM + FBS (10%), several hours prior to co-incubating, the cells were washed with PBS and DMEM without FBS was added. We therefore could analyse if the RAW 264.7 cells were being phagocytosed and if the protein was present in the cytosol.

Figure 9-Well 1: 100:1 MOI . Well 2: 300:1 MOI. Well 3. 1000:1 MOI

The above experiment illustrated that the 300:1 MOI gave the highest IRFP signal, this would suggest that the higher number of L.lactis is inducing a cell stress that results in apoptosis. Cell viability studies would be necessary to determine the extent of the cell death and compare the amount of viable cells between the two conditions. However, this experiment suggests that the protein is inside the cytosol of the macrophage cells and hence that phagosomal escape has occurred. The only source of the co-factor is now the cytosol of the macrophage cells. The presence of the co-factor in the FBS would be at a higher concentration than that in the cytosol of the macrophages, therefore the intensity readout is reduced.
L.lactis alone in DMEM and FBS was also shown to produce fluorescence whereas L.lactis alone in DMEM without FBS didn’t produce fluorescence confirming that IRFP is only active in the presence of the cofactor.

Figure 10-L. lactis only without FBS. Well 1. 100:1. Well 2. 300:1. Well 3. 1000:1
In the absence of FBS, the IRFP expressed by L.lactis wasn’t fluorescent due to the absence of biliverdin. This confirms what we would have expected initially from the literature on IRFP.
In contrast if FBS is included in the media, the presence of the co-factor allows for fluorescence.

Figure 11-L. lactis in DMEM + FBS. Well 1. 100:1 MOI. Well 2 300:1 MOI. Well 3. 1000:1 MOI.

Our results suggest that there is a linear relationship between the number of bacteria expressing IRFP in media + FBS and the number of bacteria that was initially added. A greater amount of bacteria would express a greater amount of protein and so this relationship would be predicted. As far we know, this is the first account of bacterial expression of IRFP producing fluorescence.
Filonov, G. S., Piatkevich, K. D., Ting, L.-M., Zhang, J., Kim, K. and Verkhusha, V. V. 'Bright and stable near-infrared fluorescent protein for in vivo imaging'.
Sedlak T.W, Snyder S.H. Bilirubin Benefits: Cellular Protection by a Biliverdin Reductase Antioxidant Cycle

