| (8 intermediate revisions by 2 users not shown) | |||
| Line 12: | Line 12: | ||
</div> | </div> | ||
<div class="contentRow"> | <div class="contentRow"> | ||
| − | <div class="contentCell | + | <div class="contentCell"> |
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/a/ac/T--Tuebingen--pNZ124_wiki.png" style="cursor: zoom-in;"> | <img src="https://static.igem.org/mediawiki/2016/a/ac/T--Tuebingen--pNZ124_wiki.png" style="cursor: zoom-in;"> | ||
| − | <figcaption> </figcaption> | + | <figcaption>Picture of an agarose gel electrophoresis loaded with our pNZ124-T vector. The band is higher than expected but we were able to amplify the vector in <em>E. coli</em>. </figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
<div class="contentCell"> | <div class="contentCell"> | ||
| − | <p>To introduce our targets for overexpression into L. johnsonii we planned to use the pNZ124 shuttle vector. We were able to ligate it from IDT part sequences and expressed it succesfully in E. coli. Before cloning the enzymes into the vector we introduced a terminator general terminator sequence right after the multiple cloning site (MCS). In Figure 1 a linearized terminator containing vector (pNZ124-T) can be seen. The expected length of the pNZ124-T is 2866 bp.</p> | + | <p>To introduce our targets for overexpression into L. johnsonii we planned to use the pNZ124 shuttle vector. We were able to ligate it from IDT part sequences and expressed it succesfully in E. coli. Before cloning the enzymes into the vector we introduced a terminator general terminator sequence right after the multiple cloning site (MCS). In Figure 1 a linearized terminator containing vector (pNZ124-T) can be seen. The expected length of the pNZ124-T is 2866 bp. In Figure 1 its band is higher than expected but it seems to be a suitable vector as it provides an antibiotic resistance to <em>E. coli</em>.</p> |
<p>We were able to confirm some of our parts via analytical digest followed by an agarose gel electrophoresis (Figure 2). Some of those parts were additionally approved by sequencing. These parts were Phosphofructokinase and Ketohexokinase, as well as the parts PezT, the zinc fingers ZF ZFN-L, ZF K230R, ZF-C7C7 and the composite part Kill1 of our side project Clone Wars. | <p>We were able to confirm some of our parts via analytical digest followed by an agarose gel electrophoresis (Figure 2). Some of those parts were additionally approved by sequencing. These parts were Phosphofructokinase and Ketohexokinase, as well as the parts PezT, the zinc fingers ZF ZFN-L, ZF K230R, ZF-C7C7 and the composite part Kill1 of our side project Clone Wars. | ||
</p> | </p> | ||
</div> | </div> | ||
| + | </div> | ||
| + | <div class="contentRow"> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/3/32/T--Tuebingen--Gesamtgel.png" style="cursor: zoom-in;"> | ||
| + | <figcaption> Summary of all constructs which were successfully cloned.</figcaption> | ||
| + | </figure> | ||
</div> | </div> | ||
<div class="contentRow"> | <div class="contentRow"> | ||
| Line 28: | Line 34: | ||
</div> | </div> | ||
<div class="contentRow"> | <div class="contentRow"> | ||
| − | <div class="contentCell"> | + | <div class="contentCell" style="width: 50%;"> |
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/b/bc/Tuebingen_Fructosetest.jpeg" /> | <img src="https://static.igem.org/mediawiki/2016/b/bc/Tuebingen_Fructosetest.jpeg" /> | ||
| Line 37: | Line 43: | ||
</div> | </div> | ||
<div class="contentCell"> | <div class="contentCell"> | ||
| − | <p> | + | <p>To prove the affinity of our assay for fructose we had to test various parameters. We were interested in the general sensitivity of the indole reaction. So we generated a calibration curve from fructose in water. This was our starting point. |
| + | But since the MRS Broth contains a huge amount of glucose that not necessarily will be metabolized there is the potential problem of interference due to chemical similarity. | ||
| + | <p>To disprove this we also made several conjugations of glucose and fructose in water in different concentrations and measured them. It seems like in water there would be a sufficient accuracy in our assay up to a 40 fold excess of glucose. But we were not able to produce reliable results for fructose measurement in glucose containing MRS broth. </p> | ||
</div> | </div> | ||
</div> | </div> | ||
| + | <div class="contentRow"> | ||
| + | <h1>Discussion</h1> | ||
| + | </div> | ||
| + | <div content="contentRow"> | ||
| + | <div class="contentCell contentNarrowCell"> | ||
| + | |||
| + | </div> | ||
| + | <div class="contentCell contentMirijamsRaumZumAtmen"></div> | ||
| + | <div class=contentCell"> | ||
| + | <p>Regarding all planned parts, the Cas construct for lactobacilli and the sucrose transporter could not be cloned due to difficult restriction sites. Therefore no knockout experiments were performed. The phosphofructokinase and fructose transporter were assembled to be tested for expression in lactobacilli. | ||
| + | <p>In the course of the project, we were not able to transform any constructs into <em>L. johnsonii</em>. Lactobacilli survived the transformation protocol, indicated by growth in MRS medium after electroporation. All cells died in liquid medium and on plates after incubation with antibiotics. This lead to the assumption that the pNZ124Tue vector either wasn’t taken up by the cells or the chloramphenicol-acyltransferase wasn’t expressed by the cells. | ||
| + | For cloning purposes pNZ124Tue was successfully amplified in E. coli proving that the vector works as shuttle in E. coli. We were not able to prove that pNZ124Tue can be used as expression vector for lactobacillus johnsonii. | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 00:34, 20 October 2016


Results
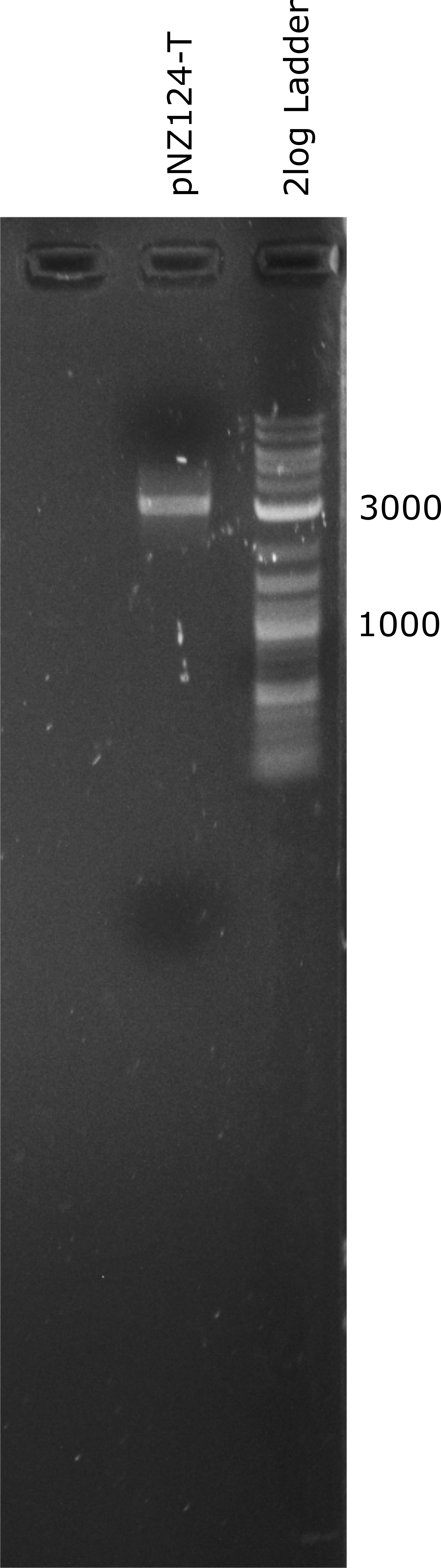
To introduce our targets for overexpression into L. johnsonii we planned to use the pNZ124 shuttle vector. We were able to ligate it from IDT part sequences and expressed it succesfully in E. coli. Before cloning the enzymes into the vector we introduced a terminator general terminator sequence right after the multiple cloning site (MCS). In Figure 1 a linearized terminator containing vector (pNZ124-T) can be seen. The expected length of the pNZ124-T is 2866 bp. In Figure 1 its band is higher than expected but it seems to be a suitable vector as it provides an antibiotic resistance to E. coli.
We were able to confirm some of our parts via analytical digest followed by an agarose gel electrophoresis (Figure 2). Some of those parts were additionally approved by sequencing. These parts were Phosphofructokinase and Ketohexokinase, as well as the parts PezT, the zinc fingers ZF ZFN-L, ZF K230R, ZF-C7C7 and the composite part Kill1 of our side project Clone Wars.

Fructose test

To prove the affinity of our assay for fructose we had to test various parameters. We were interested in the general sensitivity of the indole reaction. So we generated a calibration curve from fructose in water. This was our starting point. But since the MRS Broth contains a huge amount of glucose that not necessarily will be metabolized there is the potential problem of interference due to chemical similarity.
To disprove this we also made several conjugations of glucose and fructose in water in different concentrations and measured them. It seems like in water there would be a sufficient accuracy in our assay up to a 40 fold excess of glucose. But we were not able to produce reliable results for fructose measurement in glucose containing MRS broth.
Discussion
Regarding all planned parts, the Cas construct for lactobacilli and the sucrose transporter could not be cloned due to difficult restriction sites. Therefore no knockout experiments were performed. The phosphofructokinase and fructose transporter were assembled to be tested for expression in lactobacilli.
In the course of the project, we were not able to transform any constructs into L. johnsonii. Lactobacilli survived the transformation protocol, indicated by growth in MRS medium after electroporation. All cells died in liquid medium and on plates after incubation with antibiotics. This lead to the assumption that the pNZ124Tue vector either wasn’t taken up by the cells or the chloramphenicol-acyltransferase wasn’t expressed by the cells. For cloning purposes pNZ124Tue was successfully amplified in E. coli proving that the vector works as shuttle in E. coli. We were not able to prove that pNZ124Tue can be used as expression vector for lactobacillus johnsonii.

