(Protein Purification 2/2) |
(Excluded Protein Purification) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 43: | Line 43: | ||
| − | === Protocol === | + | === Protocol Part === |
To acquire electrocompetent ''Bacillus subtilis'', the bacteria had to be cultivated overnight in LB | To acquire electrocompetent ''Bacillus subtilis'', the bacteria had to be cultivated overnight in LB | ||
medium. 500 μl of the preculture were added to each of the 3 flasks containing 20ml of competency medium (LB | medium. 500 μl of the preculture were added to each of the 3 flasks containing 20ml of competency medium (LB | ||
| Line 121: | Line 121: | ||
== Protein Purification == | == Protein Purification == | ||
| − | + | See what we planned to do to analyze the expression of our proteins <br /> | |
| − | + | <html><a class="btn btn-default" href="//2016.igem.org/Team:UBonn_HBRS/Description/B_Subtilis/Protein_Purification">Protein Purification</a></html> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 01:16, 20 October 2016
B. Subtilis
General
Bacillus subtilis is a gram-positive bacterium which can be found almost everywhere, especially in soil. It can form a protective endospore to resist extreme environmental conditions.
We decided to work with this organism, because Bacillus subtilis is a bacteria, which naturally secretes proteins
and enzymes into its surrounding medium.
The goal was to utilise this attribute for enzyme production and create a “cell factory”. Instead of
lysating bacteria, it was desired to have a continuous production of enzymes in a liquid culture that
we can extract from the supernatant.
Repeated inoculations of new bacteria cultures are not necessary, because they will stay in the
stationary phase. Only the media has to be replaced.
Bacillus subtilis is classified as a GRAS-organism (general regarded as safe) by the U. S. Food &
Drug administration and widely used in S1 laboratories as a model organism. It is also present in
economy as probiotics in pharmacies or fungicide in agriculture.
Furthermore biotechnological applications already use Bacillus subtilis to synthesize enzymes.
Usage
For using our product we currently have two different approaches.
Either just using the supernatant to deink our paper fibers or make the time intensive and expensive
step of purifying our enzymes and then proceed with deinking the paper. For purification we
attached a His-tag to our enzymes to separate them from other components of the supernatant
with nickel columns.
In order to reach the project goals plasmids containing the needed genes had to be transformed into
the bacteria.
We worked with two different Bacillus subtilis strains which were donated by the department of
Pharmaceutical Biotechnology of the Greifswald University, Germany.
We got the wild type strain ATCC 6051 as well as the promising LS8P-D strain. The LS8P-D stain
is a derivate from the protease-deficient WB800 strain, where two additionally genes were deleted.
The lack of proteases is essential to secrete our enzymes without damaging them.
After trying out multiple protocols, we decided to accomplish transformation by electroporation.
The principle of electroporation relies on creating small holes in the cell wall of bacteria, by
sending a current through them. The plasmids enter the bacteria through the holes before the
organism repairs them.
The iGEM Team Freiburg and the iGEM Team Bonn collaborated in order to obtain working transformation protocols for Bacillus subtilis. The resulting manual “How to Bacillus subtilis” can be found on our site:
Protocol Part
To acquire electrocompetent Bacillus subtilis, the bacteria had to be cultivated overnight in LB medium. 500 μl of the preculture were added to each of the 3 flasks containing 20ml of competency medium (LB medium containing 0.5M Sorbitol) and the cultures were incubated at 37°C 250 rpm shaking till a OD600nm between 0.5 and 0.7/ml was reached. Glycine was added as a weakening agent, replacing the alanine in the cell wall leading to loosen it, for total concentrations of 1%, 1,25% and 1,5% in the different flasks. This was followed by a 60min incubation. The cells were cooled on ice for 15min and were pelleted by centrifuging at 8500 rpm for 10min at 4°C. The supernatant was removed and the cell pellet was washed repeatedly through a circle of resuspending, pelleting and discarding of supernatant. The washing steps were performed with washing buffer (0.5M Sorbitol, 0.5M Mannitol and 10% Glycerol). Finally all pellets were combined and resuspended in washing buffer. Aliquots were made and were frozen in liquid nitrogen before being stored at -80°C.
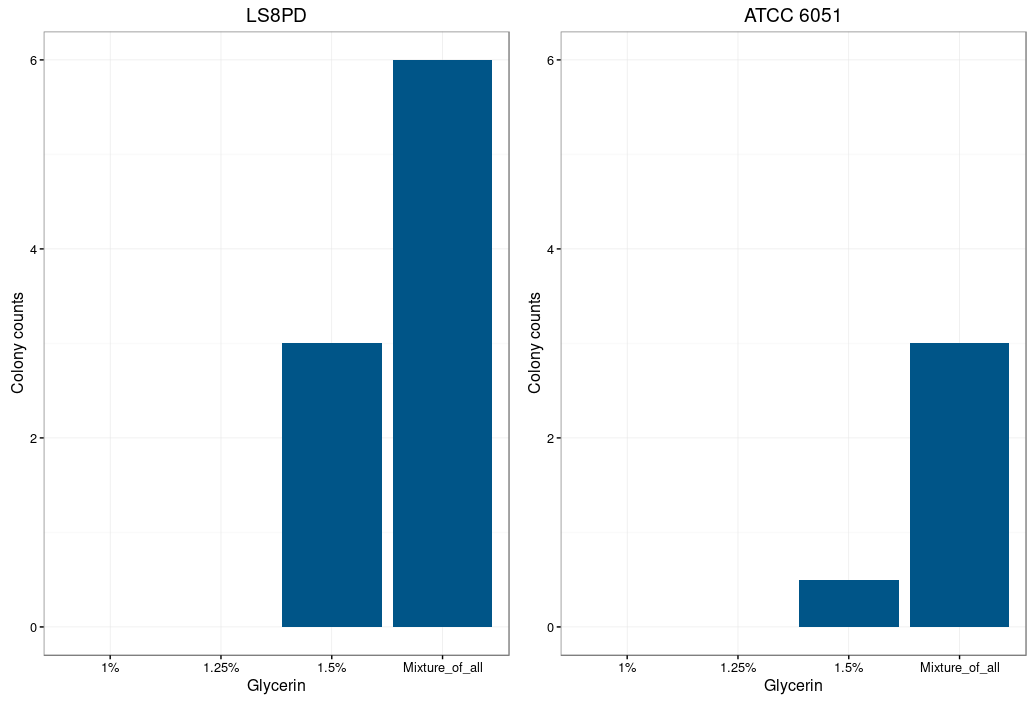
Glycine is supposed to act as a weakening agent, by replacing the alanine in the cell wall and thereby loosening it 2.
Electroporation worked best with a higher glycine concentration. However a too high concentration prevents bacterial growth and could also be toxic. The highest colony number was measured by using a mixture of all three glycine concentrations (1%,1.25% and 1.5%) for the preparation of electrocompetent cells.
In order to transform the microorganisms, an electroporator was used with the fitting cuvettes.
For the electroporation a mix out of plasmid DNA and competent bacteria was prepared with a volume of
60 μl containing 10ng of DNA per μl. The mixture was cooled on ice for 10 min along with the
electroporation cuvette and was electroporated at 2,1 kV. 1 ml of competency medium was
used to flush the cells out of the cuvette and the culture was incubated at 37°C 250 rpm shacking for
about 3h before it was plated on LB agar containing kanamycin.
| DNA-concentration (ng/μl) | Number of colonies |
|---|---|
| 10 | 11 |
| 5 | 3 |
| 1 | 1 |
| 0 | 0 |
The table shows the number of formed Bacillus subtilis colonies after transformation using electroporation with varying DNA-concentrations. It shows that a higher amount of DNA (10 ng/μl) leads to a higher number of colonies
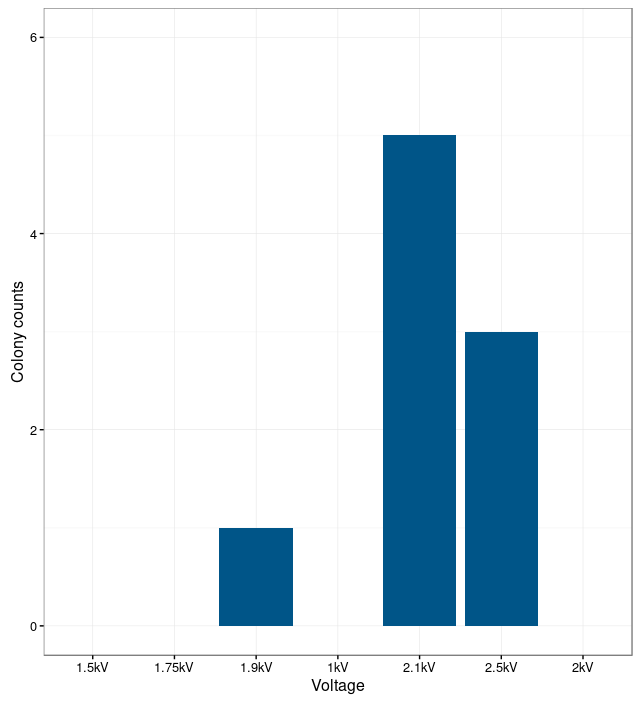
In order to find the optimal electroporation current, electroporation was performed with different voltages. The electroporation was unsuccessful at low currents. The bacteria were not able to take up the plasmids. With too high currents, the bacteria were lysed. The results show that the optimal current for electroporation is at 2.1 kV. At this current the number of successfully transformed bacteria was the highest.
Protein Purification
See what we planned to do to analyze the expression of our proteins
Protein Purification
1: http://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/
2: Zhi Zhang et al.: Development of an Efficient Electroporation Method for Iturin A-Producing Bacillus subtilis ZK


