| Line 11: | Line 11: | ||
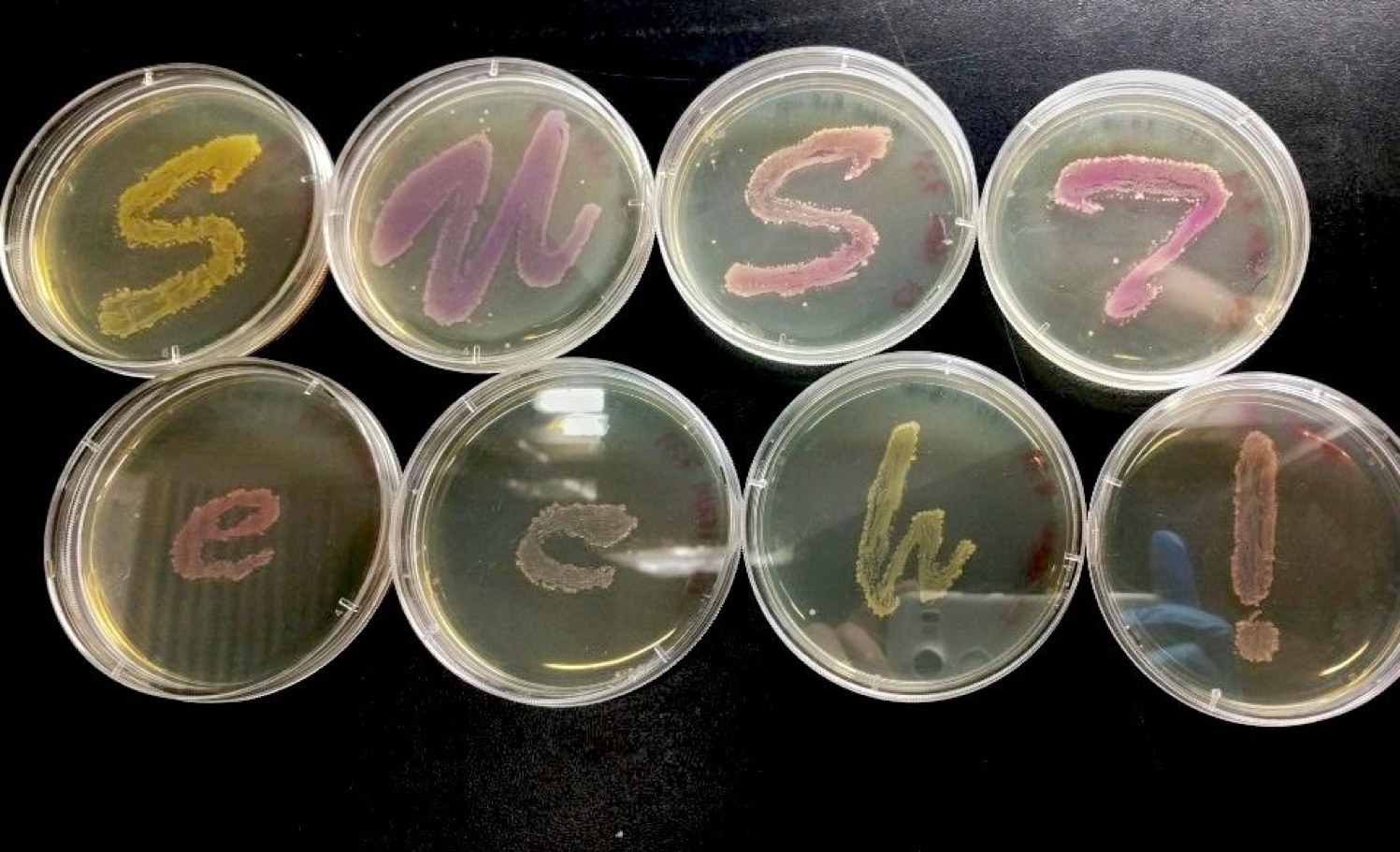
We successfully constructed 8 plasmids with various pigment coding sequence, including eforRed, gfasPurple, Fw Yellow and SpisPink. | We successfully constructed 8 plasmids with various pigment coding sequence, including eforRed, gfasPurple, Fw Yellow and SpisPink. | ||
| − | |||
{| class="table table-striped" | {| class="table table-striped" | ||
Revision as of 09:35, 18 October 2016

Composite Parts
Contribution
1. Chromoprotein collection
We successfully constructed 8 plasmids with various pigment coding sequence, including eforRed, gfasPurple, Fw Yellow and SpisPink.
| Name | Description | Length | Feature |
|---|---|---|---|
| <partinfo>BBa_K1943000</partinfo> | fwYellow, yellow chromoprotein reporter system (Strong Promoter, Strong RBS) | 912bp | |
| <partinfo>BBa_K1943001</partinfo> | fwYellow, yellow chromoprotein reporter system (medium Promoter , weak RBS) | 914bp | 221x67px |
| <partinfo>BBa_K1943002</partinfo> | gfasPurple, purple chromoprotein reporter system (Strong Promoter, Strong RBS) | 867bp | 224x58px |
| <partinfo>BBa_K1943003</partinfo> | gfasPurple, purple chromoprotein reporter system (medium Promoter , weak RBS) | 869bp | 227x58px |
| <partinfo>BBa_K1943004</partinfo> | eforRed, Red chromoprotein reporter system (Strong Promoter, Strong RBS) | 879bp | 204x58px |
| <partinfo>BBa_K1943005</partinfo> | eforRed, Red chromoprotein reporter system (medium Promoter , weak RBS) | 881bp | 217x62px |
| <partinfo>BBa_K1943006</partinfo> | spisPink, Pink chromoprotein reporter system (Strong Promoter, Strong RBS) | 876bp | 214x57px |
| <partinfo>BBa_K1943007</partinfo> | spisPink, Pink chromoprotein reporter system (medium Promoter , weak RBS) | 878bp | 212x55px |
Compared to the common used fluorescence protein such as GFP, RFP, chromoproteins are obviously more useful reporter genes. In our parts, the coding sequences of these pigments are all among 700bp long, and the expression efficiency is high according to our experiment results.
We contributed this series of the chromoprotein plasmid for the reason that they have significant advantages to be used as backbones. The length of 700bp is easy to be distinguished from the original backbone (around 2000bp) by gel electrophoresis when constructing new expression system. Additionally, this kind of backbones are superior because we are able to directly observe the expression results by using naked eyes. It will be much more convenient for the laboratory with limited conditions and also save our time. In the tests, the color of colonies were easy to recognize after 18 hours of incubation.
Our plasmid can also be a substitute for J04450, which kindly recommended to be used in plasmid construction by replacing the RFP gene. For the same reason, we provide these pigment sequence to make the construction process more efficiently.
Source
The following list is the Biobricks we’ve used in construction and their detailed design information.
| Part | Type | Description | Backbone | Location on the plate |
|---|---|---|---|---|
| <partinfo>BBa_K880005</partinfo> | Promoter + RBS | Strong Promoter + Strong RBS | pSB1C3 | 3F 2016 Kit Plate 2 |
| <partinfo>BBa_K608007</partinfo> | Promoter + RBS | Medium Promoter + Weak RBS | pSB1C3 | 5G 2016 Kit Plate 1 |
| <partinfo>BBa_K592012</partinfo> | Chromoprotein | eforRed, red chromoprotein | pSB1C3 | 15I 2016 Kit Plate 6 |
| <partinfo>BBa_K1033919</partinfo> | Chromoprotein | gfasPurple, purplr chromoprotein | pSB1C3 | 9K 2016 Kit Plate 6 |
| <partinfo>BBa_K1033910</partinfo> | Chromoprotein | fwYellow, yellow chromoprotein | pSB1C3 | 6K 2016 Kit Plate 4 |
| <partinfo>BBa_K1033932</partinfo> | Chromoprotein | spisPink, pink chromoprotein | pSB1C3 | 11K 2016 Kit Plate 6 |
| <partinfo>BBa_B0015</partinfo> | Terminator | Double terminator | pSB1C3 | 3F 2016 Kit Plate 3 |
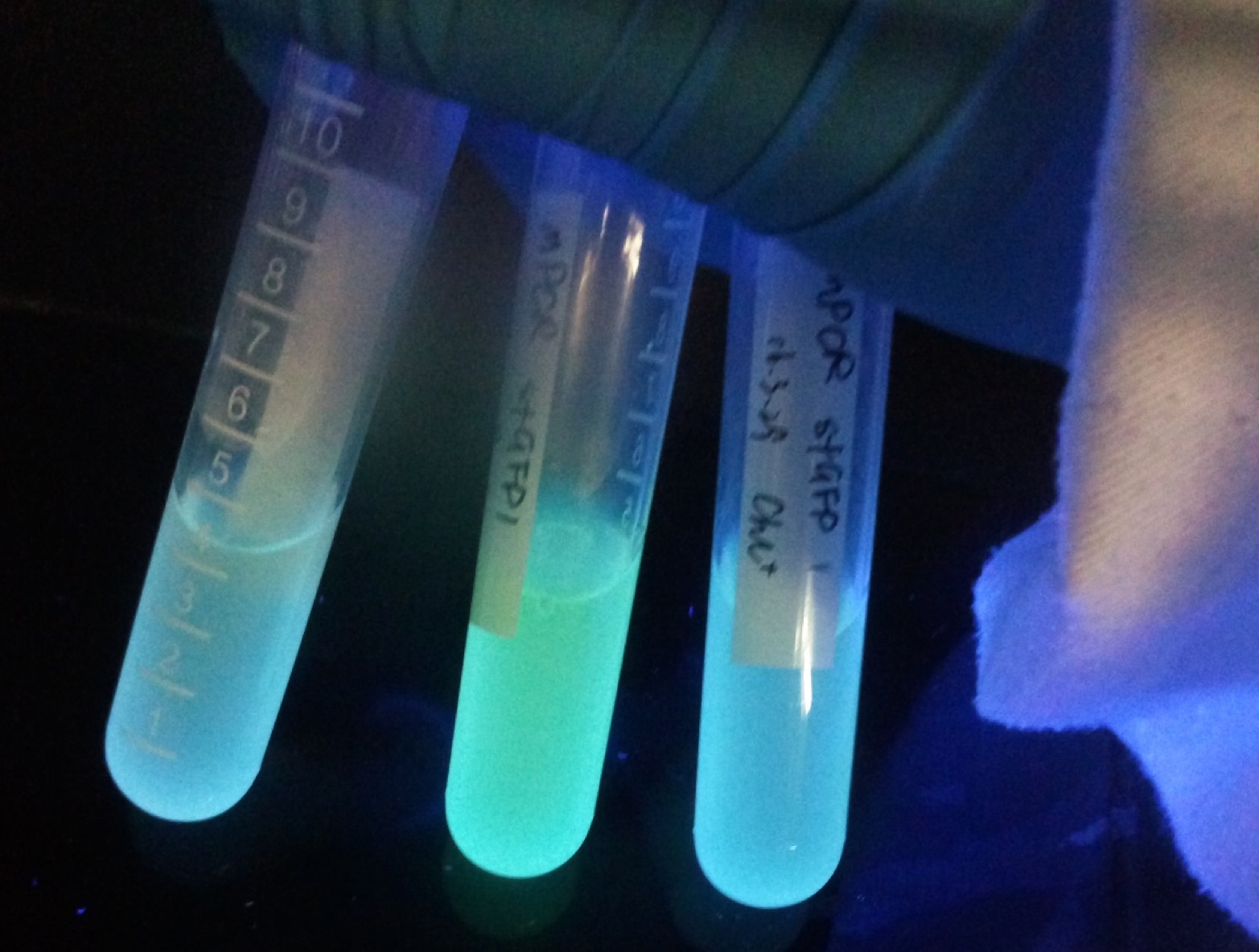
2. sfGFP collection
We submitted this part of super folder GFP (sfGFP) with a strong promoter J23116. Under this expression system, a very bright blue-green fluorescence will be observed after 6 hours of incubation. Its excited wavelength is close to the UV light. We commonly manipulate it by using the UV in super clean bench. The fluorescence is strong enough to be observed by naked eyes. For this reason, sfGFP is always used as reporter gene under the plasmid construction.
This plasmid can be used as a vector. After ligation, it is easy to pick up the single colony without blue-green fluorescence.
On the other part we submitted mutated sfGFP (msfGFP). It has one base-pair difference with the original sfGFP on its 165th site. We changed the base pair from AT to GC by using whole plasmid mutagenesis to remove KpnI restriction site. We kindly hope the little effort can bring convenience to the following iGEMers.
In addition, we also constructed the sfGFP expression system with a terminator following the coding sequence. While the two groups (with and without terminator) showed no significant difference in our experience. We hope these parts can be provided as comparable experimental groups to quantify the efficiency of bacterial terminators.
| Name | Description | Length | Feature |
|---|---|---|---|
| <partinfo>BBa_K1943008</partinfo> | Strong promoter+ strong RBS+ sfGFP | 778bp | 133x52px |
| <partinfo>BBa_K1943009</partinfo> | Strong promoter+ strong RBS+ sfGFP+ terminator | 918bp | 206x49px |
| <partinfo>BBa_K1943010</partinfo> | Strong promoter+ strong RBS+ msfGFP | 781bp | 145x50px |
| <partinfo>BBa_K1943011</partinfo> | Strong promoter+ strong RBS+ msfGFP+ terminator | 915bp | 215x50px |