Proof of Concept
Project
Contents
Overview
To implement audio-genetics, we chose two (Piezo1 and TRPC5) from hundreds of mechanosensitive(MS) channels that are relevant to sensing mechanical stress such as sound wave, because they are more sensitive to mechanical stress and more selectively permeable to calcium ion compared with others. Additionally, we used calcium indicator (R-GECO) to quantitatively examine the sensibility of Piezo1 and TRPC5 with the help of microfluidics and hypo-osmolality, the former one was to exert shear force on our transgenic CHO-K1 cell's membrane, the latter one was to exert tension on cell's membrane.
We also examined the intracellular calcium response to sound wave with different combination of frequency and intensity in advance, then we quantitatively analyzed the regulation ability of audio-genetics in long term by using special promoter (pNFAT) with its downstream GFP gene.
Finally, to ensure a required sensitivity to mechanical stress and select a series of audio respondent channels, we used protein engineering by directed evolution to get mutated sTRPC5 (super TRPC5) with high sensitivity to mechanical stress. All in all, our project is well organized. Each procedure is indispensable. The whole project can strongly prove the possibility of using sound wave energy as the extracellular input signal to induce intracellular downstream signal.
1. Testing the PiggyBac System
In order to quantitatively test the function of PiggyBac system, we made two groups of transfection using Lipofectamine® 3000. One was a group of CHO-K1 cells transfected with R-GECO, and the other group was the cells transfected with R-GECO and PiggyBac.
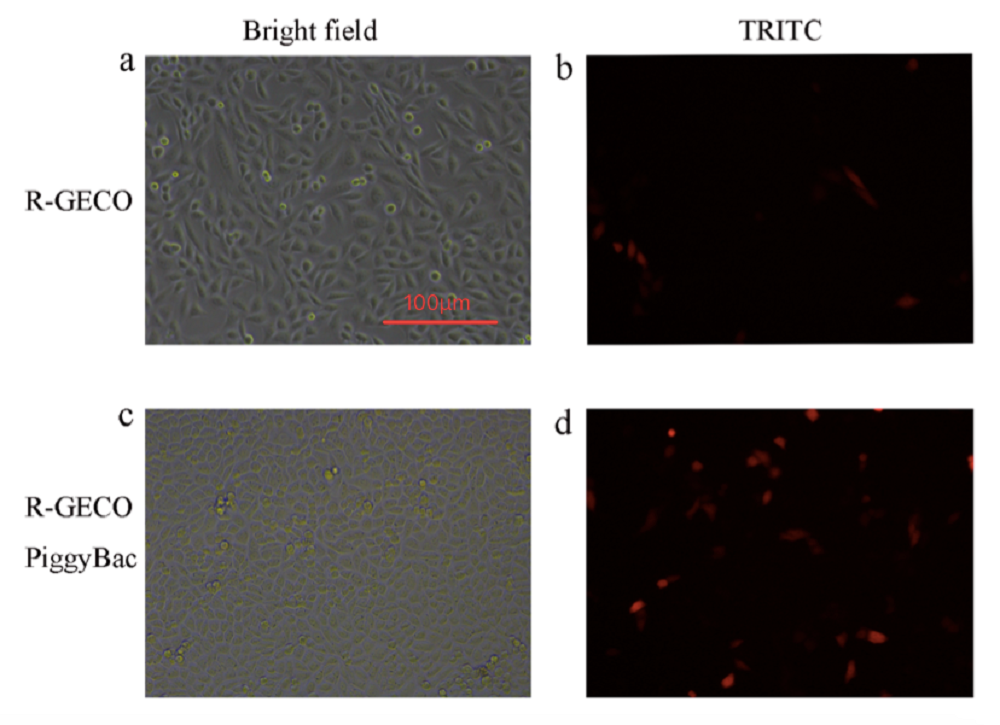
Comparing the fluorescence image of the cell transfected with R-GECO (Fig. 1B) with R-GECO + PiggyBac (Fig. 1D) on the second day after transfection, we can observe that the group with PiggyBac has more fluorescent cells. It proves that PiggyBac transposon system can help the insertion of R-GECO gene into CHO-K1 genomes.
2. Examination of R-GECO Function
To quantitatively test the function of R-GECO, two experiments, potassium chloride (KCl) induced calcium influx and ionomycin induced calcium influx, were conducted under the live cell imaging.
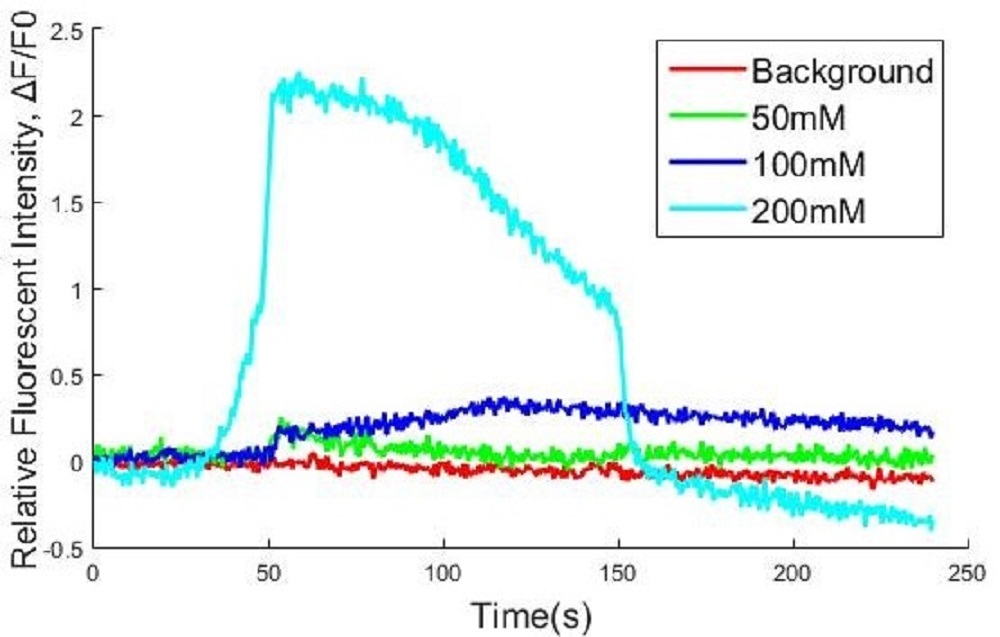
Calcium influx due to depolarization of cell membrane was induced by different concentration of KCl. Different volume of KCl was added by drops into cell culture wells to reach the different final concentrations of KCl (0mM control, 12.5mM, 50mM, 200mM, respectively) at 40 second.
There was no distinct fluorescent increasing at 12.5mM KCl comparing with the control group. The cells under 50mM and 200mM KCl conditions both emitted high fluorescence, and the fluorescence of cells at 200mM was higher than cell at 50mM along with oscillation. It was proved quantitatively that R-GECO could be excited by KCl induced calcium influx.
A B
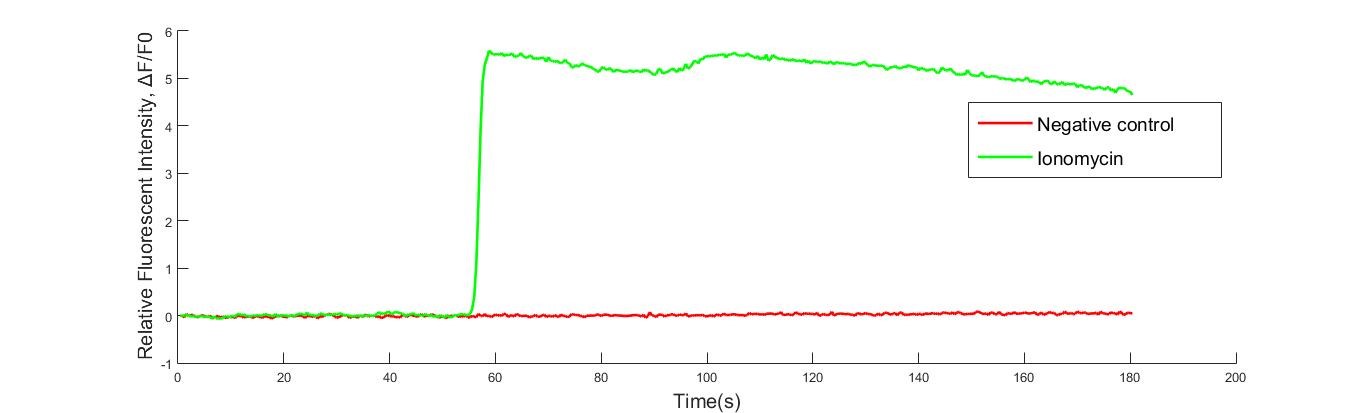
Fig. Ionomycin induced Ca2+ influx result. Fluorescence intensity over time is shown as signal per pixel (the initial intensity has been normalized to 0). The data are shown as mean ± SEM, n = 5 for control, 10 μM ionomycin.
R-GECO’s response to calcium influx was also tested by ionomycin which was added at 20s. After adding ionomycin (10μM final concentration), the increase of R-GECO fluorescence was distinct. Because the ionomycin was dropped beside the observed region of cells, the direction of fluorescence gradient (shown in the movie) was in accordance with the direction of ionomycin diffusion.
3. Measure Piezo1 and TRPC5 Sensitivity
We designed two experiments to quantitatively examine the sensibility of Piezo1 and TRPC5. Microfluidics and hypoosmolality were employed to exert shear force and tension on our transgenic CHO-K1 cell's membrane separately.
Microfluidics
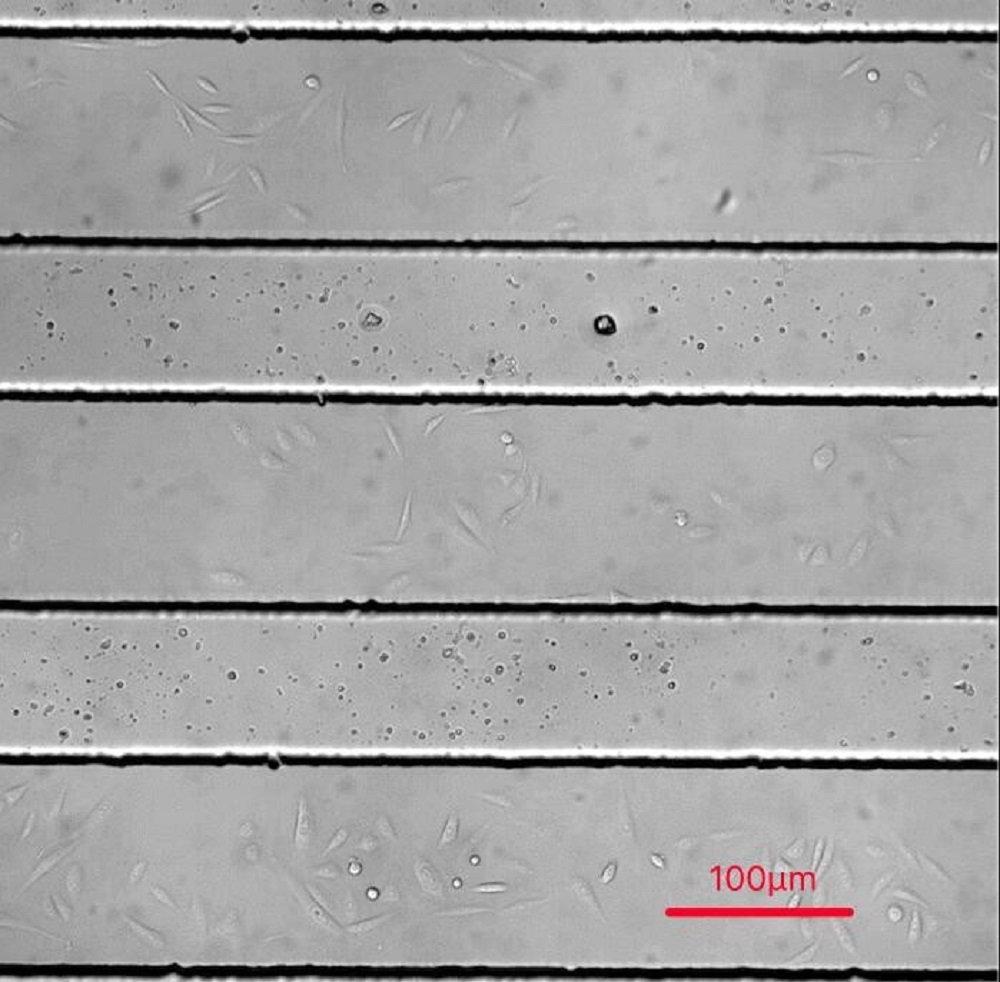
Fig.1 a Schematic structure of microfluidics channel
CHO-K1 cells were cultured adherent on the bottom glass of PDMS tunnel. Stable and flexible force field was formed surrounding cell, directly controlled by the pumped-inflow rate.
See the design of microfluidics chip and the simulation of force field at https://2016.igem.org/Team:SUSTech_Shenzhen/Model
CHO-K1 cell suspension was injected into gelatin treated microfluidics chips and incubated at 37°C, 5% CO2 incubator for 24 hours. Then the culture medium was injected into the chip channels (with the inflow rate of 25μL/min, 50μL/min and100μL/min controlled by micro syringe pump). The pump of culture medium started at t=10s. Cells’ response towards different shear force was measured by R-GECO fluorescence intensity under live cell imaging. (waiting result)
discussion! (modeling)
Hypoosmolarity
A
B
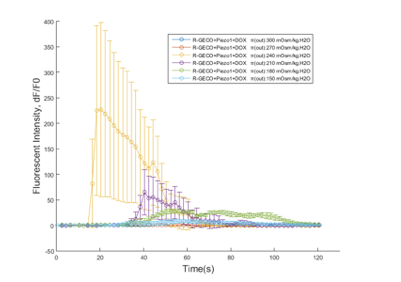
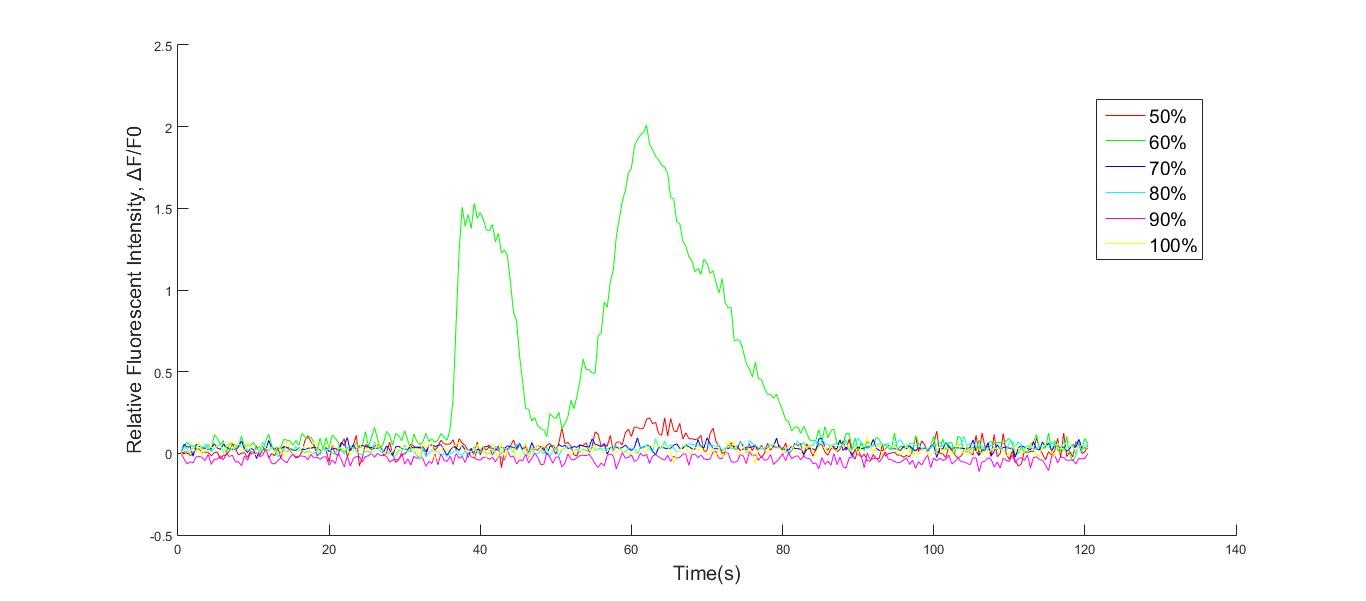
Fig. Osmotic pressure test result. Fluorescence intensity over time is shown as signal per pixel (the initial intensity has been normalized to 0). The data are shown as mean ± SEM, n = 5 for 3 groups of 100%, 90%, 80%, 70%, 60% or 50%.
Calcium influx was indicated by R-GECO fluorescence intensity. Culture medium was replaced by HHBSS before using live cell imaging. Three groups of cells were seeded into 18 wells: 6 wells of R-GECO, 6 wells of Piezo1 (without doxycycline inducement) and 6 wells of Piezo1 (with doxycycline inducement). Distilled water (added with CaCl2) was added into wells to make concentration of HHBSS into 100%, 90%, 80%, 70%, 60% or 50% for 3 groups.
4. Analyze the regulatory ability of audiogenetics
(高大山 高大山 )
5. Select through Cre-loxp system
Cre-loxp system played an important role in directed evolution, because we needed to screen a single cell colony only with one loxp site in the genome of each CHO-K1 cell for single copy insert of mutated TRPC5. Here we tested the function of cre-loxp system by making 3 different groups of transfection in the cells that already inserted with loxp gene on genomes using Lipofectamine® 3000.
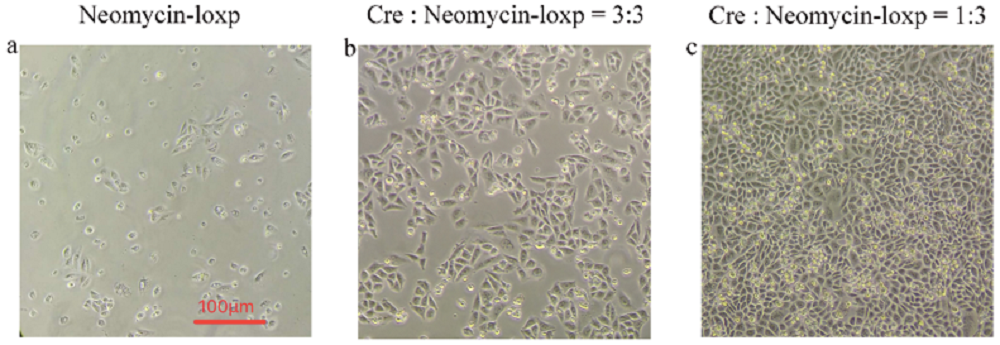
Few cells survives (Fig1. a) after antibiotics screening without transfecting Cre recombinase gene, which means that there is a small possibility that loxp gene can insert into genomes by interior transposase in cells. To find out the optimal ratio of Cre recombinase gene to Neomycin-loxp gene in the process of transfection, we compared two different ratio, 3:3 (Fig1. b) and 1:3 (Fig1. c), with equal amount of Neomycin-loxp gene but different amount of Cre recombinase gene.
From observation, we know that excess Cre recombinase is not appropriate in transfection process. The most possible reason is that surplus Cre recombinase is harmful for cells, so they may random cut and recombine genes in cell’s genome. Thus, we finally chose 1:3 as the ratio of Cre recombinase gene to Neomycin-loxp gene in transfection.
6. Mutagenesis Library of TRPC5
Mutation frequency calibration
1. Define ideal mutation frequency
Mutation frequency is the product of DNA polymerase error rate and number of duplications, it depends on initial target amount and fold amplification. Mutation frequencies of 1~4 amino acid changes (2~7 nucleotide changes) per gene are commonly employed.
We have mutated the Ankyrin repeats (734bp length) of TRPC5 by employ 500ng initial tartget and 32 cycle amplification according to the protocol of GeneMorph II Random Mutagenesis Kit (Agilent Technologies,Catalog #200550).
2. Calibration of mutation frequency by sequencing
To calibrate the mutation frequency in experimental condition, we sequenced 20 mutated TRPC5 samples in the region of ankyrin repeats. The table below shows the correspondence between mutation(s) and sequenced sample numbers.
| Mutation(s) | 1 | 2 | 0 |
|---|---|---|---|
| Sample Numbers | 4 | 1 | 14 |
Table 1 Practical mutations and sample numbers
5 samples have been detected mutation(s). However, the transformation result showed that the digestion might be incomplete (Table 2). Still, the data could offer us some information. Basically, the practical mutation frequency with initial target amount of 500ng was similar with the theoretical mutation frequency.
| Colonies | Control |
|---|---|
| 10688 | 113 |
Table 2. Transformation result of mutated TRPC5 ligation product vs control
To construct a large-scale mutation library with different mutation frequencies in the specific region, we also did error-prone PCR with different initial target amounts to get TRPC5 mutants with low, medium and high mutation rate.
Library construction
3. Calculate competent cell efficiency from experimental result
(1). Formula: \frac{\text{Colonies}}{\text{ng of DNA plated}}\times 1000ng/\mu g
(2). Calculation: 237 / 0.02 x 1000=1.185x107 cfu/μg
(3). Actual meaning: 20pg, 5640bp TRPC5 plasmid → 3.457x106 copies
3.457x106 / 237=14586, which means 1 of 14586 plasmids would actually grow on the plate.
4. Compare three methods for mutation library construction
| Method | Colonies | Control |
|---|---|---|
| R. E. D and Ligation | 1772 | 330 |
| Whole Plasmid Mutagenesis | 191 | 0 |
| Gibson Assembly | 38 | 7 |
Table 5. Transformation Results of Three Methods for Mutation Library Construction. All groups were transformed with 5μl products after all experiment procedure with 100ul competent cell. Restriction Enzyme Digestion and Ligation method and Gibson assembly method used the same mole of TRPC5 backbone. Transformation operations were all the same.
From the table, we can know Restriction Enzyme Digestion and Ligation method is the most efficient way of constructing library.
(1). 62ng, 4906bp plasmid → 1.232x1010 copies
(2). Efficiency= \frac{(1772-330)\times 20 \div 5}{1.232 \times 10^10 \times \frac{1}{14586}}\times 100\% = 0.68\%
We needed to consider their effect on CHO-k1 cell if being directly transfected without transformation and plasmid construction. Since the ligation efficiency was 0.68%, we did an experiment comparing the survival rate of CHO-k1 cells after being transfected with linear DNA and circular DNA. After antibiotic screening, we luckily found that the former one were almost dead, while the latter one were good. Thus we could directly transfect the ligation products into cell without transformation, preventing the mutation library from decreasing.
7. Screening cells for Audio-genetics test
1. Transfect the cells with loxp gene, pNFAT+GFP gene, R-GECO gene (calcium indicator) by PiggyBac system and sort them into single colonies by FACS (BD FACSAria)
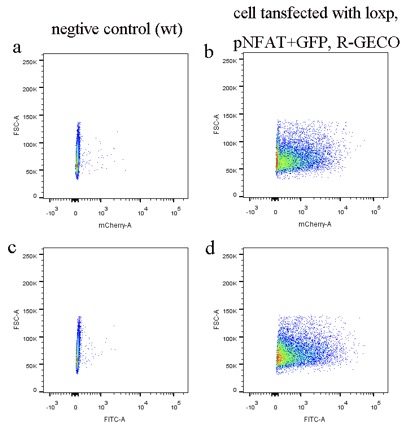
The data was collected in the process of sorting by FACS, total cells count is 10000. Cells transfected with R-GECO gene will continually express R-GECO, because CAG promoter is a constitutive promoter. Thus, most cells have red fluorescence (Fig1. b) compare with wild type control (Fig1. a). Cells transfected with pNFAT+GFP should not express GFP because promoter NFAT need to be activated by calcium signal indirectly. However, most cells still can express GFP (Fig. d) compare with wild type control (Fig. c), which mean there is some leak expression of promoter NFAT. 2. Select the single cell colony only with one copy number of loxp by qPCR To evaluate the efficiency of each mutated channels, single cell colony with only one copy number is required when testing, so we designed the following experiments to achieve it. 2.1 Primer design and primer efficiency test
The real-time PCR primers (Forward: TTGTCAAGACCGACCTGTCC, Reverse: TTCAGTGACAACGTCGAGCA) were designed by Primer-BLAST, a tool for finding specific primers, followed by BLAST specificity check. Firstly, we selected an appropriate annealing temperature to avoid unnecessary amplification as well as improving primer efficiency by normal PCR test in wild type CHO-K1 genomes, the result indicates that 62℃ may be the most appropriate annealing temperature. In addition, we also confirmed that the amplification product was accurate at 62℃ when the target was cells genome that transfected with neomycin-loxp gene by sequencing the PCR product.
Real-time PCR ( here we used TaKaRa-SYBR Premix Ex Taq II(Tli RNaseH Plus) ran in the ABI StepOne Plus qPCR instrument) can be used to calculate the primer efficiency of PCR amplification via Equation 1. In which Xn = PCR product quantity after n cycles, X0 = initial copy number, E = amplification efficiency, n = cycle number.
Xn=x0 \times (1+E)^n
Equation 1
The efficiency of the primer is 104% by calculating (Within the usual margin of error, which is 90%~110%). Also, the melt curve has only one peak, which means the designed primer is quite specific to the transfected CHO-K1 cell’s genomic DNA.
2.2 Select the cell colony with single copy number
In each qPCR screening experiment, we made a standard curve by employing a series of 5-fold dilution gradient Neomycin-loxp gene with known mole amount. Therefore, we could quantitatively examined the copy number of neomycin-loxp gene in single colony genomes by fitting the sample Ct data in the standard amplification plot. Standard curve was made by using the following data.
| Relative Quantity | log(Relative Quantity) | Ct |
|---|---|---|
| 1 | 0 | 15.01047802 |
| 0.2 | -0.698970004 | 16.94664574 |
| 0.04 | -1.397940009 | 19.24198341 |
| 0.00032 | -3.494850022 | 25.79216194 |
| -4.193820026 | 27.9509449 |
Table 10. Log(Relative Quantity) vs. Ct
Figure 15 Fig. 15 Fitted Standard Curve
(within the usual margins of error, which is 90%~110%)
Fig16
Figure 16. Melt Curve of Neomycin-loxp Transfected CHO-K1 Cell’s gDNA
Copy Number
| Mass/ng | Ct | Relative Quantity | 10^-3 fmole | Copy Number |
| 78.8075 | 23.2236 | 0.002110101 | 0.0531425 | 4.054 |
| 40.7625 | 24.1791 | 0.001040142 | 0.0274875 | 3.863 |
| 19.0225 | 25.2316 | 0.000477161 | 0.0128275 | 3.797 |
| Average | 3.905 |
Table 11. Neomycin-loxp Copy Number of CHO-K1 Cell
Relative Quantity = 10 \frac{\text{Ct}-14.902}{-3.1101}
Copy Number :
Plasmid used in standard curve: 0.1021 fmol, Relative Quantity=1
fomular (c: copy number)
fig13 Figure 13. Amplification Plot for Standard
fig14 Figure 14. Amplification Plot for Sample
The efficiency of PiggyBac transposon system ranged from 2% for one transposon to 0.5% for five transposons [1]. Consequently, it’s quite possible for us to screen out the cells with single or low copies of Neomycin-loxp.
In practical experiments, we totally test 18 different single colony genomes. The lowest copy number of those selected CHO-K1 cell transfected with Neomycin-loxp is around 4, which was not good enough. The reason may be that the designed primer was not so satisfying. Also, the contamination during real-time PCR was hard to avoid. To get cells with single copy of Neomycin-loxp, we still need to design better primers as well as paying attention to the experiment operations.
3. Transfect the cells with mutated TRPC5 by Cre-loxp system
We are doing these experiments.
use fluorescence activated cell sorting (FACS) to select the cells with high intensity of GFP after long term stimulate
We are doing.
[3] Lu, X. and W. Huang, PiggyBac mediated multiplex gene transfer in mouse embryonic stem cell. PLoS One, 2014. 9(12): p. e115072.
- ↑ Lu, X. and W. Huang, PiggyBac mediated multiplex gene transfer in mouse embryonic stem cell. PLoS One, 2014. 9(12): p. ell 5072