
Contents
- 1 Abstract
- 2 Introduction
- 3 Experiments & Results
- 3-1 Primary Stage: We successfully enhanced upon the surface display system INPNC in E. coli
- 3-2 Secondary Stage: We successfully observed the binding of NoVLP to surface expressed scFv expressing E. coli
- 3-3 Tertiary Stage: We succeeded in observing the binding of CBD to cellulose
- 3-4 Quaternary Stage: We tried dual expression of the two surface expressing proteins
- 4 Discussion
1 Abstract
Norovirus (NoV) accounts for 18% of the diarrheal disease in the world, with more than 200 thousand people dying each year from NoV infection [1]. Despite this overwhelming damage caused by NoV infection, the world has yet to develop a direct approach to combat them. Here we, iGEMKyoto, propose 'Noro-catcher', a new biodevice that binds to and ultimately expels NoV from human intestine. This biodevice consists of two types of functional handles that are each fused to surface expressing domains, both expressed in the outer membrane of E. coli.
The first handle is the anti-NoV scFv (single-chain variable fragment). With this handle, we aim our Noro-catcher to bind to NoV within various environments, e.g. sewage, water supplies, test tubes in virus detection assay, and small intestines in our body.
The second handle is the cellulose binding domain (CBD). With this handle, we aim our Noro-catcher to be "leashed" to cellulose as opposed to it being "free roaming" within human digestive tracts. We ultimately aim to use our Noro-catcher for therapeutic purposes, removing E. coli-bound NoV from human intestine with our sterilized Noro-catcher. The CBD handle can aid in swift removal of our biodevice by linking them to cellulose, as cellulose passes human intestine undigested.
To express these two handles in the outer membrane of E. coli, we enhanced a surface expression protein domain called INPNC, and used it as the anchor. By scanning electron microscopy, we observed the binding of E. coli expressing INPNC-His-scFv bound to NoV-like particles, which is the capsid proteins of NoV. By fluorescent microscopy, we have also observed the binding of cellulose and E. coli, which expressed INPNC-His-CBDcex to cellulose.
We demonstrated each of the Noro-catcher handles' functionality by physically binding them to NoVLP and cellulose. As shown by our project, creating a recombinant bacteria expressing combination of target-binding modules dramatically enhances each modules' potentials. This mechanism can be applied to removal system of other harmful agents, such as other pathogens, organic toxins, and heavy metals.
2 Introduction
2-1 Global burdens of NoV infection and progress of its countermeasures
Norovirus (NoV) causes inflammation of the stomach and/or intestines, which is called gastroenteritis. When infected with NoV, a person usually develops symptoms in 12 to 48 hours, and most will get better within 3 days [2][3]. However, when its symptoms worsen, it can lead to death. Its common symptoms include diarrhea, vomiting, nausea, and severe stomach pain. Fig1, 2 show the recent prevalence of NoV infection among the world.
The data for U.S. and U.K. seem relatively low (Fig1), but NoV cannot be ignored in those developed countries. As shown in Fig2, NoV remains a significant issue to overcome in both developed and developing countries.

The data indicates more than 10 in 1 people of Africa are infected by NoV annually [4][5][6].

Almost half of all food poisonings are caused by NoV in the two countries [5][6].
There already exist treatments and drugs to alleviate individual symptoms like vomiting and nausea. However, there are none to target NoV itself. As NoV's infection mechanisms and its cultivation condition was not known until recently, research on such methods has been significantly hindered [*1]. We therein resolved to devote our knowledge and skills of synthetic biology to join the global fight against the unconquered and highly contagious NoV.
Recent studies show that NoV seems to be multiplying in the human B cells, binding to the Histo-blood group antigen (HBGA) of enteric cells to get within the intestinal epithelial barrier [3][5][7]. Another study shows that human anti-NoV antibodies, such as antibody 12A2, bind to NoV's HBGA-binding site [8]. It suggests that the antibody sterically hinders NoV from binding to HBGA and blocks NoV's entry into human body, effectively neutralizing them (Fig3).
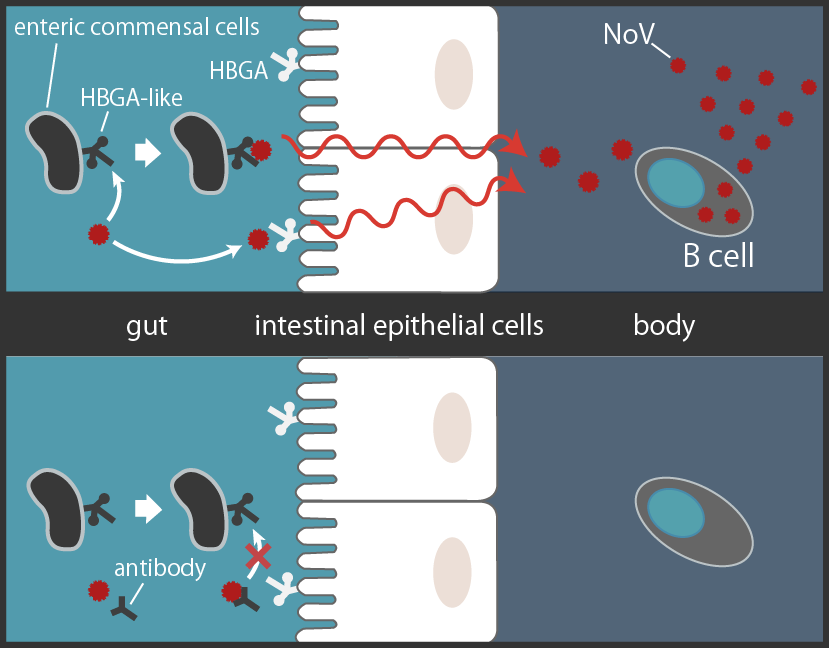
NoV binds to the HBGA to pass through intestinal epithelial cells to multiply in the B cells. Human antibody 12A2 sterically hinders NoV's binding to HBGA[*2] [3][5][7][8][9].
2-2 Development of Noro-catcher
From these studies stated above, we thought of using a well known bacteria, E. coli to be a carrier of anti-NoV antibody to intestinal tracks. If we succeeded in delivering the anti-NoV antibody that binds to HBGA binding site of NoV, not only would it block the binding of NoV to HGBA, but might also physically remove E. coli-bound NoV from the intestinal tracks.
There are three steps in which we aim our Noro-catcher to carry out its intended purpose (Fig4). First, we deliver our Noro-catcher into the patient intestine. Second, after reaching the NoV-infested intestine, Noro-catcher will bind specifically to NoV using its surface expressed anti-NoV antibody. Third, the NoV-bound Noro-catcher will promptly be excreted from human digestive system. To achieve the first step, we could use exiting methods such as orally administering medicine, sterilized and capsule-enveloped, to patients. In our study, we created a biodevice with two handles in order to achieve the second and third steps. These two handles are both achieved through the same surface expression technology.

1) Noro-catcher is sterilized and enveloped within a capsule, then orally administered to patients.
2) Noro-catcher binds to NoVs multiplying within the small intestine.
3) Binding Noro-catcher to indigestible cellulose for swift removal of the NoV-bound Noro-catcher.
2-3 Surface Display
For this Noro-catcher to function, it is essential to express a functional antibody and CBD on the bacterial cell surface. We thus undertook the method commonly called surface display (Fig5).

Fusing passenger protein to anchoring protein anchored in lipid bilayer (the outer membrane of E. coli) with a linker peptide. Passenger will physically be anchored to the cell surface of E. coli.
Surface display is a method to fix passenger proteins to the cell surface using anchoring domain. We selected INPNC and BclA proteins as the anchor to fix passenger proteins. INPNC is a domain of a surface expressing protein derived from Pseudomonas Syringae [6]. BclA is a hair-like protein derived from Bacillus anthracis(34F2) that covers the B. anthracis' spores. Despite its surprisingly compact size of 21 amino acid long chain, BclA is known to display passenger proteins on bacterial outer membrane [10]. When INPNC and BclA's original proteins are surface expressed, their N-terminal domains (NTD) faces the inner cell, C-terminal domains (CTD) faces outwards, and internal repeating domains fill the gap between NTD and CTD.
Although most anchoring domains are limited in its ability to carry large passenger proteins, INPNC and BclA are unique in that they can carry up to 120 kDa [10]. As the groundwork for our Noro-catcher, we considered the highly versatile INPNC and BclA as suitable anchors.
2-4 Anti-NoV Antibody and NoVLP
We used single-chain variable fragment (scFv) of anti-NoV antibody. scFv is a fusion protein consisting of VH and VL, a variable region of the antibody. VH and VL is fused by a linker peptide. (shown in Fig6).

To test scFv's functionality, we decided to use NoV-like particles (NoVLP) instead of NoV particles, which are highly infectious (<20 copies to be infected) and too dangerous to be used in our lab. NoVLP consists only of capsid proteins of NoV, and does not include the viral RNA. By scanning electron microscopy (SEM), it was observed that NoVLP takes a similar form to NoV. We also confirmed that NoVLP is recognized as the antigen by both the original antibody and scFv. Therefore, we expected that we can perform a highly accurate simulation of our Noro-catcher's activity against NoV, by using NoVLP.
2-5 Cellulose Binding Domain
Among numerous CBDs with various characteristics and binding affinity, we decided to use CBDcex for our Noro-catcher, as it was found on iGEM Registry thanks to iGEM Bielefeld 2012. CBDcex binds irreversibly to cellulose [11].
CBDcex derives from the exoglucanase of a bacteria, Cellulomonas fimi. This bacteria is known for high cellulolytic activities, using cellulose as a source of energy. CBDcex (10.3 kDa) is a 100 amino acid long domain of an exo-beta-1,4-glucanase (47.1 kDa) [12]. CBDcex belongs to the Carbohydrate Binding Module family 2.
2-6 Development Stage for Noro-catcher's Functionality
In order to prove its practicality and functionality, we set four stages that our Noro-catcher should clear.
- Establish bacterial surface display system for both scFv and CBD in E. coli
- Observe the functionality of surface expressed scFv by its binding to NoVLP
- Observe the functionality of surface expressed CBD by its binding to cellulose
- Dual express surface expressed scFv and CBD in E. coli and observe the Noro-catcher's functionality
We organized our results in accordance to the above four stages.
3 Experiments & Results
3-1 Primary Stage: We successfully enhanced upon the surface display system INPNC in E. coli
1) Construction of enhanced parts for surface displayAs a device for surface display system, INPNC and BclA are already made into BioBrick parts by past iGEM teams. For example, WPI-Worcester 2014 surface expressed YFP (yellow fluorescent protein) using BclA and observed it through fluorescence microscopy. Penn 2012 used immunofluorescence microscopy to observe surface expression of INPNC-HA tag fusion protein. The functionality of their parts are well described in their respective pages.
We started our projects by modifying these existing parts to enhance their functions. As shown in Fig7, our plasmids encode 6xhistidine tag (His tag) directly downstream of the INPNC and the BclA domain. The His tag polypeptide can be detected in Western blotting in similar manner as HA tag in the INPNC-HA system. In addition, His tag has a unique property of enabling fusion protein purification under denaturing conditions. The fusion proteins used for surface display will be expressed in the insoluble membrane fraction of E. coli. Therefore, this His tag was expected to enable additional options in the characterization of the fusion proteins [*3].
2) Construction of scFv plasmids and CBD plasmidsThe scFv we used binds specifically to NoV GII.4 strain [8]. We have kindly been given the scFv-encoding plasmid (NoV GII.4 strain-specific antibody 12A2 (vector: pSCCA5-E8d)) from Dr. Kyoko Moriguchi (Fujita Health University). We used the scFv coding region of this plasmid.
For the CBD, we chose a BioBrick part K1321342 from iGEM Imperial 2014 for the construction of CBDcex [*4]. We placed the passenger protein (scFv or CBDcex) encoding region downstream of the INPNC-His and BclA-His surface expression modules. His tag was placed in between the two domains. The resulting constructs were sequenced and verified (Fig7).

The passenger protein (scFv or CBDcex) encoding region were placed downstream of the INPNC-His and BclA-His surface expression modules. His tag was placed in between the two domains.
3) INPNC-His and BclA-His can be detected in Western blotting with a single anti-His tag antibody regardless of the passenger protein placed downstream.
Among the parts submitted in the past, Penn 2012 used HA tag to detect INPNC-(passenger protein)-HA tag fusion protein. We anticipated our His tag plasmids can be used for protein detection in the similar manner. However, we noticed on their wiki page that they did not test whether their INPNC-HA parts can be detected through HA tag if passenger proteins are placed downstream. We tested whether our His tag can be used for the full length detection of fusion proteins with passenger proteins. We used whole cell Western blotting using anti-His tag antibody for this purpose.

Marker, Negative Control(BBa_K165002), BclA-His-scFv(33 kDa), INPNC-His-scFv(63 kDa), BclA-His-CBDcex(17 kDa), INPNC-His-CBDcex(47 kDa), Positive Control(BBa_K875004)(43 kDa)
Whole cell lysates were prepared from E. coli DH5alpha strain carrying various plasmids shown in the figure. Samples were separated by 10% SDS-PAGE and transferred to a PVDF membrane. His tag proteins were visualized by anti-His tag monoclonal antibody (clone #OGHis, MBL, Japan).
Bands corresponding to INPNC-His-scFv (63 kDa) and INPNC-His-CBDcex (47 kDa) in their respective lanes were observed, which confirms expression of the fusion proteins. We also observed that BclA-His-scFv band (lane 3) was much smaller than the expected size (33 kDa). This may suggest that the protein coded by this plasmid may be prone to intracellular cleavage. From these results, we concluded our INPNC-His-(passenger protein) and BclA-His-(passenger protein) can be detected with anti-His tag antibody, and we eliminated the need to use individual antibodies for passenger proteins when using our INPNC-His or BclA-His surface expression modules.
4) INPNC-His and BclA-His can be purified using nickel columns that specifically bind to His tag, regardless of the passenger proteins placed downstreamWe then examined if the proteins' His tag can be used for pulldown under denaturing conditions by nickel beads. We tested this property by purifying INPNC-His-(passenger protein) and BclA-His-(passenger protein) from the membrane fraction of the cell, using Nickel Sepharose beads (GE) affinity purification. We prepared the membrane fraction from the E. coli lysate by ultracentrifugation, and solubilized them using 7M guanidine-HCl, then purified the target protein using Nickel Sepharose that specifically binds to his-tag (Fig9).


Membrane fraction was solubilized and used for Nickel Sepharose purification and precipitates were examined by Western blotting using anti-His tag antibody. The bands corresponding with INPNC-His-scFv (63 kDa) INPNC-His-CBDcex (47 kDa) were observed in their respective lanes, while the bands corresponding with BclA could not be observed. Marker, Negative Control (BBa_K165002), BclA-His-CBDcex (17 kDa), INPNC-His-CBDcex(47 kDa), BclA-His-scFv (33 kDa), INPNC-His-scFv (63 kDa), Positive Control (BBa_K875004, 43 kDa)
The bands appearing in lane BclA-His-scFv and INPNC-His-scFv at 43 kDa are spillover from the Positive Control.
We concluded that INPNC-His modules can be detected and purified using His tag, even with the passenger protein fused directly downstream. This is a significant enhancement to the INPNC parts submitted by Penn2012, BBa_K811005. Thus we registered and submitted this new part to the iGEM Registry, BBa_K1933001. The results shown above also confirm INPNC-His-(passenger protein)'s expression, and suggest they are expressed on the cell surface as expected.
3-2 Secondary Stage: We successfully observed the binding of NoVLP to surface expressed scFv expressing E. coli
1. Sample preparation and Scanning Electron Microscopy (SEM)In 3-1 we established a system of surface expression using INPNC-His module. The essential part of Noro-catcher lies in its binding to the NoV using surface expressed scFv. Thus, we decided to test scFv's ability using NoVLP. We first conducted a Western blotting for NoVLP using anti-NoVLP antibodies (Fig11). The NoVLP and anti-NoVLP antibodies were kindly given to us by Dr. Daisuke Sano (Hokkaido University).

Samples were separated by 10-20% gradient SDS-PAGE and transferred to a nitrocellulose membrane. Anti-NoVLP polyclonal antibodies (rabbit) was used for the detection. A single clear band was observed at 58 kDa. 1. Protein size marker, 2. NoVLP
We observed a single band at 58 kDa. This corresponds to the molecular weight of the monomer of capsid protein VP1, which makes up NoVLP [13]. There were no clear indication of small peptides that could mean occurrence of degradation. Thus we used this for the next assay.
We then tested the functionality of scFv to bind to NoV. To this end, we prepared INPNC-His- and BclA-His-scFv expressing E. coli and incubated them with NoVLP. We then used low-speed centrifugation to recollect E. coli together with bound NoVLP, and removed the supernatants which should contain unbound NoVLP. We observed the cells with SEM (Fig12).

1, 2. E. coli expressing BclA-His-scFv. 3-6. E. coli expressing INPNC-His-scFv, 7,8. Sample with only NoVLP. 2-microm scale bars are shown in each picture.
As shown clearly in the pictures, we observed a characteristic spherical objects in extremely close proximity to the cell surface of the INPNC-His-scFv expressing E. coli. We could not observe a similar phenomenon with BclA-His-scFv, presumably due to the cleavage in the cell (Fig8, lane BclA-His-scFv) and absence in the membrane fraction (Fig10 lane 4, see signals at 33 kDa). This spherical objects match size with the reported size of the recombinant NoVLP, which is from 40 to 100 nm [14]. Furthermore, as it matches in form with the spherical objects observed in samples only with NoVLP, we concluded this spherical object as the NoVLP, and concluded that the INPNC-His-scFv expressing E. coli bound to NoVLP successfully.
2. The functionality of the surface display system was visually demonstratedThis observation suggests that our primary objective of binding NoV to E. coli using surface expressed scFv was achieved. In addition, this result further solidifies the functionality of INPNC-His surface display module in E. coli. This visual demonstration of the functionality of INPNC surface expression modules encourages further application of this part by future iGEM teams. We added this observation to Penn2012's parts experience page.
3. INPNC-expressing E. coli was observed with characteristic deformityWhen comparing SEM pictures, we noticed that E. coli using INPNC system seems to be more elongated compared to those with BclA system. To confirm this, we determined aspect ratios of all E. coli from the SEM pictures using ImageJ (see Materials and Methods for more detail ). By Welch's t-test (two-sided), we found that E. coli with INPNC system were in fact more elongated than BclA system expressing E. coli (p=0.0015 <0.05).
We have also made a growth curve of the INPNC-His- and BclA-His- system expressing E. coli. (Fig13). It shows that INPNC-His-scFv expressing E. coli has significant delays in growth compared to its BclA counterpart. The results from the growth curve and elongated shapes both suggest that the expression of INPNC-His-scFv to the outer membrane caused significant stress and deformation to the host E. coli.

Measurements were done for 10 hours, but the graph only shows 4 hours of measurement since the medium saturated within 4 hours. We used E. coli harboring BBa_E1010 as the control.
3-3 Tertiary Stage: We succeeded in observing the binding of CBD to cellulose
As shown in 3-1, we observed the expression of INPNC-His-CBDcex on the membrane fraction. We next tested the passenger protein's functionality, e.g. CBDcex's binding to cellulose.
1. Observing the binding of E-coli to cellulose on a macroscopic levelCBDcex’s binding to cellulose was initially examined by measuring the optical density (OD600) of cell suspension before and after incubation with cellulose, as well as its washing buffer (Fig14), as reported previously [15]. The results were summarized in Fig15.
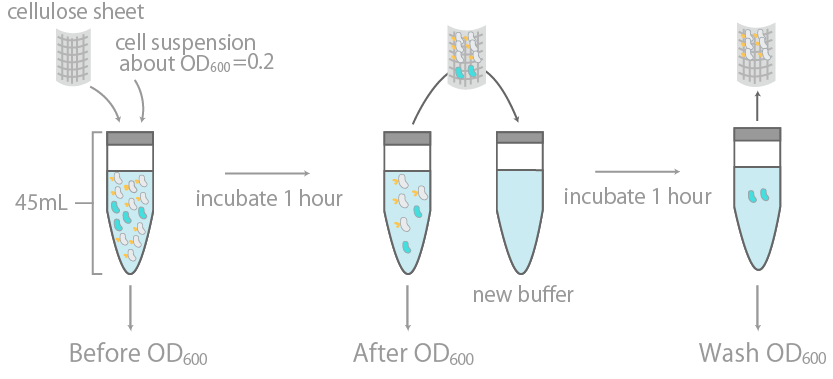
The binding efficiency was determined by measuring the optical density (OD600) of cell suspension before and after incubation with cellulose, as well as its washing buffer.

The y-axis is the OD600 of the cell suspensions. +/- indicates the presence or lack thereof cellulose during incubation. INPNC-ctl is the negative control, expressing only the INPNC domain. The linearity of OD600 and cell concentration was predetermined, with results confirming linearity in the range of about 0.04~1.00.
Unfortunately, we could not observe significant differences between the binding of INPNC-His-CBDcex to cellulose and binding of negative control (without CBDcex) to cellulose under this assay condition. This experiment was performed essentially in accordance to previously reported assays (Wang et al) [15][16]. However, there are a few differences between the two assays. They expressed CBDcex with Lpp-OmpA surface expression modules, instead of INPNC-His, to conduct cellulose binding assays. Lpp-OmpA is also a well-characterized surface display system .We concluded that our CBD's binding to cellulose under our condition was not as efficient as their reported system.
2. Observing the binding of E-coli to cellulose with fluorescence microscopyWe next conducted fluorescence microscopy to directly observe E. coli cells bound to cellulose. This would allow for detection of binding with increased sensitivity (Fig16).
Fluorescent objects are DAPI stained E. coli.
(1) Binding of E. coli to bemliese
a-1. Bemliese (5mmx5mm) only, a-2. INPNC-His-ctl (OD600=0.2, 1ml) with Bemliese(5mmx5mm), a-3. INPNC-His-CBDcex (OD600=0.2, 1ml) with Bemliese (5mmx5mm) with INPNC-His-CBDcex (OD600=0.2, 1ml).
(2) Binding of E. coli to cellulose powder
b. cellulose powder (0.5mg) only, c-1. INPNC-His-CBDctl (OD600=1.0, 1ml) with cellulose powder (0.5mg), c-2. INPNC-His-CBDcex (OD600=1.0, 1ml) with cellulose powder (0.5mg), d-1. INPNC-His-CBDctl (OD600=1.0, 1ml) with cellulose powder (0.1mg), d-2. INPNC-His-CBDcex (OD600=1.0, 1ml) with cellulose powder (0.1mg)

Sample names correspond with Sample names of Fig16.
(1) Binding of E. coli to bemliese
The results of F test and T test between a-2 and a-3; F(10,12)=5.50 ,p<0.01, t(11)=2.28, p<0.05
(2) Binding of E. coli to cellulose powder
The results of F test and T test between c-1 and c-2; F(8,9)=33.88, p<0.01, t(8)=3.24, p<0.05
The results of F test and T test between d-1 and d-2; F(10,10)=18.64, p<0.01, t(11)=3.44, p<0.01
Fig16 shows the results. After incubating E. coli with about 35~38 micrometer cellulose powder, we DAPI stained E. coli and observed it under fluorescence microscopy. As shown, CBD-surface expressing E. coli was observed binding to cellulose, showing the functional expression of INPNC-His-CBD in our system. When control plasmid was used, the numbers of cellulose-bound E. coli cells were significantly reduced (Fig17).
From these results, we concluded that our INPNC-His-CBDcex can be used to physically link E. coli and cellulose.
3-4 Quaternary Stage: We tried the dual expression of the two surface expressing proteins
We worked on the simultaneous expressions of INPNC-His-scFv and INPNC-His-CBDcex. We co-transformed the two plasmids. These two plasmids both have chloramphenicol (Cp) resistance marker, but they have different replication origins (p15A for the scFv and pMB1 derivative for the CBD), each belonging to a different incompatibility group. In theory, the two plasmids can be stably inherited. We first tested whether the Cp resistant colonies from the co-transformation have both of the plasmids with colony PCR. (Fig18)

PCR products by VF2 and VR were analyzed by electrophoresis.
As shown in Fig18, colony amplified in lane a-6 was observed to have two different plasmids. This was the first time in our project that we had both of our surface display plasmids simultaneously in an E. coli
In order to test whether the two plasmids could be stably inherited, we performed single colony isolation of this strain and conducted a colony PCR on the newly formed 4 colonies. As shown in Fig18, the band corresponding to the INPNC-His-CBDcex were observed in all the colonies, while the band for low-copy plasmid, INPNC-His-scFv couldn't be observed in any of the lanes. This result strongly suggests that the low-copy plasmid dropped out during the formation of new colonies. As both the plasmids are Cp-resistant, deleting one plasmid will not affect the E. coli's antibiotic status. In order for our Noro-catcher to stably pass down both plasmids, we need to construct one plasmid with a different antibiotic marker and grow the bacteria on a medium with two different antibiotics. In addition to changing the antibiotic resistance, it may be better to use different surface display systems for stable expression of scFv and CBDcex (see Discussion).
4 Discussion
4-1 Our Accomplishment
As stated above, we tackled on the development of Noro-catcher, a NoV infection-combatting biodevice. We constructed and expressed two handles, anti-NoV scFv and CBDcex on the bacterial surface using INPNC-His surface display system. We directly observed the binding of scFv expressing E. coli and NoVLP using SEM. Also, we observed E. coli's binding to cellulose with fluorescent microscopy, though its binding levels leave rooms for improvements. When both of the two plasmids expressing these handles were transformed into an E. coli strain, we could detect colonies carrying the two plasmids. However, the two plasmids were not stably inherited after single colony isolation, presumably due to the use of the same antibiotic resistance marker in the plasmids.
4-2 Noro-catcher can be significantly improved in the future
A major problem of our Noro-catcher is the instability in retaining two handles, scFv and CBDcex, within an E. coli cell. To solve this problem, it would be necessary to construct a stable dual-expression strain using different antibiotic resistance markers, in addition to introducing compatible plasmid pairs with different replication origins as mentioned above. It would also be an effective measure to combine the two plasmids to make a single plasmid which can express both of the handles (Fig18).
Upon constructing a dual expression system, it should be noted that there is a potential problem in expressing two different passenger proteins by using a single surface display mechanism. It was reported that the dual expression of passengers using Lpp-OmpA was unsuccessful [16]. The same display systems are likely to be expressed using the same machinery of E. coli, and are displayed on the same region of its membrane. In order to avoid the direct competition of the two proteins, it might be needed to express them in different systems. As Lpp-OmpA-CBDcex system were reported to work well [15][16][17], we can introduce this system to our project. Then, we can expect to realize a Noro-catcher which binds effectively to both cellulose and NoVLP. Our future work would be the construction of the plasmid as shown in Fig19.

4-3 Developing therapeutic application for NoV
As we successfully observed the Noro-catcher's binding to NoVLP, we have paved a new path for developing therapeutic medication for NoV infection. As stated in the introduction, NoV binding to Histo-blood group antigen (HBGA) is suggested to be an important part of NoV invasion. Also, NoV binding to antibody 12A2 in its HBGA binding site has been demonstrated previously [18]. The scFv used in our research derives from antibody 12A2, so it also sterically blocks the binding of NoV to HBGA in theory, it might neutralize NoV using the same mechanism employed by human immune system. If we can show that our Noro-catcher does indeed neutralize NoV infectivity, we will shed a new light on the underdeveloped field of NoV therapeutics.
Also, as our Noro-catcher is essentially NoV-capturing E. coli, we can cheaply mass produce them. This means that Noro-catcher can contribute to NoV therapeutics on the economical side as well. For example, the monoclonal antibodies for NoV GII.4 strains generally costs USD600~USD1000 per milliliter. Our E. coli surface expressing scFv can theoretically be multiplied and be used without limitation and with negligible costs. This may be the more economical alternative to studying antigen-antibody interactions.
4-4 Possibility of further application of our surface expression system
Our INPNC-His surface expressing system can be applied to other virus researches by interchanging the variable fragment part of scFv domains to other virus antibodies. Other applications include, but are not limited to: switching scFv domain to heavy metal binding proteins to create a bioremediation device that removes harmful metals from the environment; switching scFv domain to neutral lipid binding proteins to remove excess fat from human bodies; and so on. With our INPNC-His module, it isn't possible to just surface display passenger proteins, but also to easily identify its expression through His tag Western blotting. As such, we have also contributed to further research on bacterial surface expression systems.
Note
[*1] This summer, a team of scientists reported a way to incubate NoV using human stem cells, and studies on NoV from these fields are also expected to progress rapidly. [*2] The oligosaccharides on the surface of enteric commensal bacteria are HBGA-like substances [17]. [*3] In general, purification and characterization of proteins require it to be soluble. Insoluble samples need to be denatured and solubilized to be handled, but proteins would completely lose its characteristics and functions, disabling purification. However, His tag's unique ability to retain its property to bind to nickel after denaturing means it can exceptionally be purified even when in insoluble sample. [*4] It is iGEM Bielefeld 2012 that have made the CBDcex into Biobrick parts, and iGEM Imperial 2014 added on to these parts by fusing them with sf-GFP. Since the parts by iGEM Imperial 2014 was included in the kit, we have chosen to use theirs for the construction. For more information about the parts, please visit their respective pages.Reference
[1] Berger, Stephen. Infectious Diseases of the World. GIDEON Informatics Inc, 2015.[2] Morillo, Simone Guadagnucci, and Maria do Carmo Sampaio Tavares Timenetsky. "Norovirus: an overview." Revista da Associação Médica Brasileira 57.4 (2011): 462-467.
[3] Karst SM, Wobus CE (2015) A Working Model of How Noroviruses Infect the Intestine. PLoS Pathog11(2):e1004626.doi:10.1371/journal. Ppat.1004626
[4] Mans, Janet, et al. "Norovirus Epidemiology in Africa: A Review." PloS one 11.4 (2016): e0146280.
[5] Food Standards Agency (2012): Annual Report of the Chief Scientist https://www.food.gov.uk/sites/default/files/multimedia/pdfs/publication/csar1112.pdf
[6] Centers of Disease Control and Prevention (2016): Estimates of Foodborne Illness in the United States https://www.cdc.gov/foodborneburden/burden/index.html
[7] Ettayebi, Khalil, et al. "Replication of human noroviruses in stem cell–derived human enteroids." Science 353.6306 (2016): 1387-1393.
[8] Kyoko Higo-Moriguchi, Haruko Shirato, Yuichi Someya, Yoshikazu Kurosawa, Naokazu Takeda, and Koki Taniguchi. “Isolation of Cross-Reactive Human Monoclonal Antibodies That Prevent Binding of Human Noroviruses to Histo-Blood Group Antigens” Journal of Medical Virology 86:558–567 (2014)
[9]Miura, Takayuki, et al. "Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses." Journal of virology 87.17 (2013): 9441-9451.
[10] Park, Tae Jung, et al. "Surface display of recombinant proteins on Escherichia coli by BclA exosporium of Bacillus anthracis." Microbial cell factories 12.1 (2013): 1.
[11]Tomme, Peter, et al. "Characterization and affinity applications of cellulose-binding domains." Journal of Chromatography B: Biomedical Sciences and Applications 715.1 (1998): 283-296.
[12] MacLeod, Alasdair M., et al. "Mechanistic consequences of mutation of active site carboxylates in a retaining β-1, 4-glycanase from Cellulomonas fimi." Biochemistry 35.40 (1996): 13165-13172.
[13] Hardy, Michele E. "Norovirus protein structure and function." FEMS microbiology letters 253.1 (2005): 1-8.
[14] Tomé-Amat, Jaime, et al. "Secreted production of assembled Norovirus virus-like particles from Pichia pastoris." Microbial cell factories 13.1 (2014): 1.
[15] Wang, Aijun A., Ashok Mulchandani, and Wilfred Chen. "Specific adhesion to cellulose and hydrolysis of organophosphate nerve agents by a genetically engineered Escherichia coli strain with a surface-expressed cellulose-binding domain and organophosphorus hydrolase." Applied and environmental microbiology 68.4 (2002): 1684-1689.
[16] Wang, Aijun A., Ashok Mulchandani, and Wilfred Chen. "Whole‐Cell Immobilization Using Cell Surface‐Exposed Cellulose‐Binding Domain." Biotechnology progress 17.3 (2001): 407-411.
[17] Wang, Aijun A., Wilfred Chen, and Ashok Mulchandani. "Detoxification of organophosphate nerve agents by immobilized dual functional biocatalysts in a cellulose hollow fiber bioreactor." Biotechnology and bioengineering 91.3 (2005): 379-386.
[18] Shanker, Sreejesh, et al. "Structural basis for norovirus neutralization by an HBGA blocking human IgA antibody." Proceedings of the National Academy of Sciences (2016): 201609990.






