
The aim of our work with Clustered Regularly Interspaced Short Palindromic Repeats (CRIPSR) was to metabolically engineer the increase accumulation of our chlorophyll synthesis precursor, protoporphyrin-IX (PPIX). This was achieved by disrupting the hemH gene, there by decreasing the production of hemH, preventing the diversion of the PPIX towards heme, increasing the accumulation of PPIX. The results of this work can be seen below. Mutant colonies were detected by an increase of PPIX seen using fluorescence spectroscopy. Amongst the mutants generated, we selected the highest accumulating mutant for modelling experiments and device verification of our Mg-Chelatase Plasmid.
Summary of Results
- A functional Cas9 RNP complex was demonstrated by specifically cleaving the hemH gene PCR product as expected.
- The Cas9 RNP complex was functional in vivo as demonstrated by its effect on E. coli by killing them in a concentration dependent manner, which is expected if excess double stranded breaks are made in a RecA minus strain.
- The Cas9 mutagenesis produced a hemH mutant of E.coli which accumulates protoporphyrin IX.
Results

Metabolically Engineering with CRIPSR Results
We have designed primers to amplify the hemH gene using E.coli genomic DNA as a template. The results can be seen in the amplification products of Figure 1(a). Figure 1(b) indicates that the Cas9 RNP complex works in vitro and cleaves the PCR product into the expected sizes as designed by the target guide RNA. The expected sizes of the cleaved products are approximately 850/150bp. The incomplete cleavage of the PCR product is likely to be due to a insufficient amounts of the Cas9 guide RNA complex.
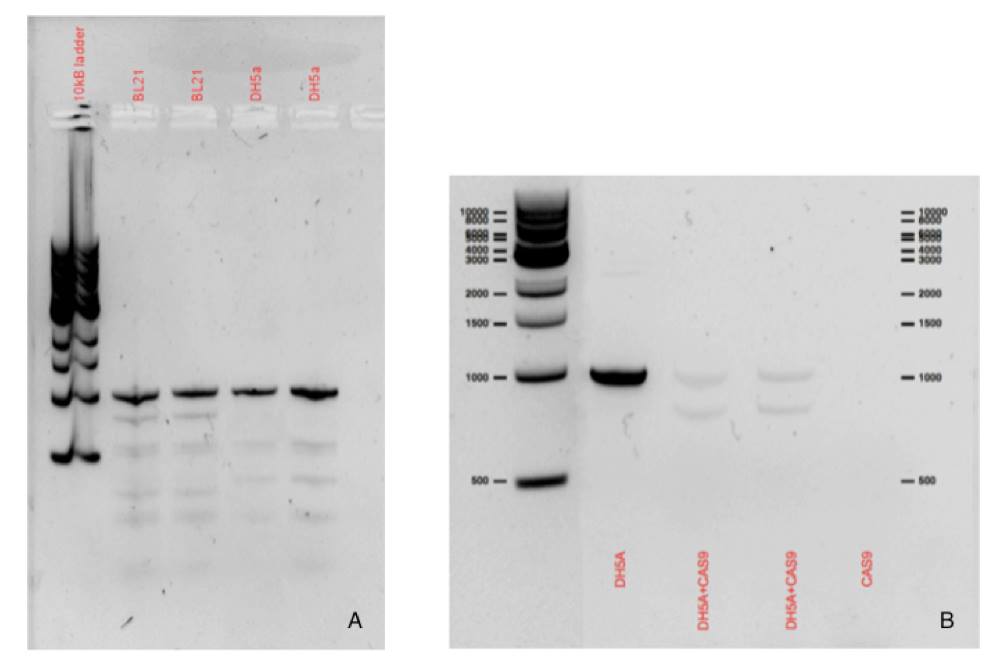
Fig 1(a). Ferrochelatase PCR products using our designed primers used to amplify the hemH gene (990bp).
Fig 1(b). In vitro assay indicating the function of Cas9. Expected sizes can be seen according to the designed target guide RNA.
The cell viability curve below outlines the survival of cells after electroporation of the Cas9 RNP
complex in vivo. Higher concentration of Cas9 RNP complex results in lower cell viability correlating with an
increase of double stranded breaks. This indicates that the Cas9 RNP complex is efficiently introduced in to the bacteria cells without a loss of
function. These results were also observed by Dutra et al [1].


Fig 2. Cell viability curve of CFU/mL against increasing Cas9 concentration. The curve indicates that increasing Cas9 concentration inversely effects cell viability.
Individual colonies from the serial dilution plates were patch plated from M9-glucose to M9-glycerol plates which is expected to cause increased production of heme to allow oxidative metabolism of the glycerol. These patched colonies were screened for the accumulation of PPIX by fluorescence spectroscopy
(Ex 404nm/ Em 600-700nm and Em 630/Ex 350-500nm). The emission peak at 630nm and excitation peak at 404nm indicates the presence of PP-IX. The excitation spectra from this screening is shown below.


Fig 3. Fluorescence spectroscopy results demonstrating the accumulation of PPIX at 404nm excitation. (A) Mutants are indicated by peaks at 620nm suggesting PPIX accumulation. (B) Control assay grown on glycerol, showing no changes in peak emission.
We found 12 colonies which were slightly coloured on the cells from Cas9 RNP complex colonies patch plated from M9-glucose to M9-glycerol plates. These were screened for fluorescence and showed PPIX accumulation. The highest accumulator, mutant #1, which was the most coloured was selected for further experiments.
Mutant #1 was streaked out to single colonies and the single colonies were screen for PPIX accumulation. The fluorescence emission spectrum (Ex 404nm) of two of these colonies is shown below, indicating significant accumulation of protoporphyrin IX within these cells. This mutant was then made competent and used in further experiments.
Mutant #1 was streaked out to single colonies and the single colonies were screen for PPIX accumulation. The fluorescence emission spectrum (Ex 404nm) of two of these colonies is shown below, indicating significant accumulation of protoporphyrin IX within these cells. This mutant was then made competent and used in further experiments.
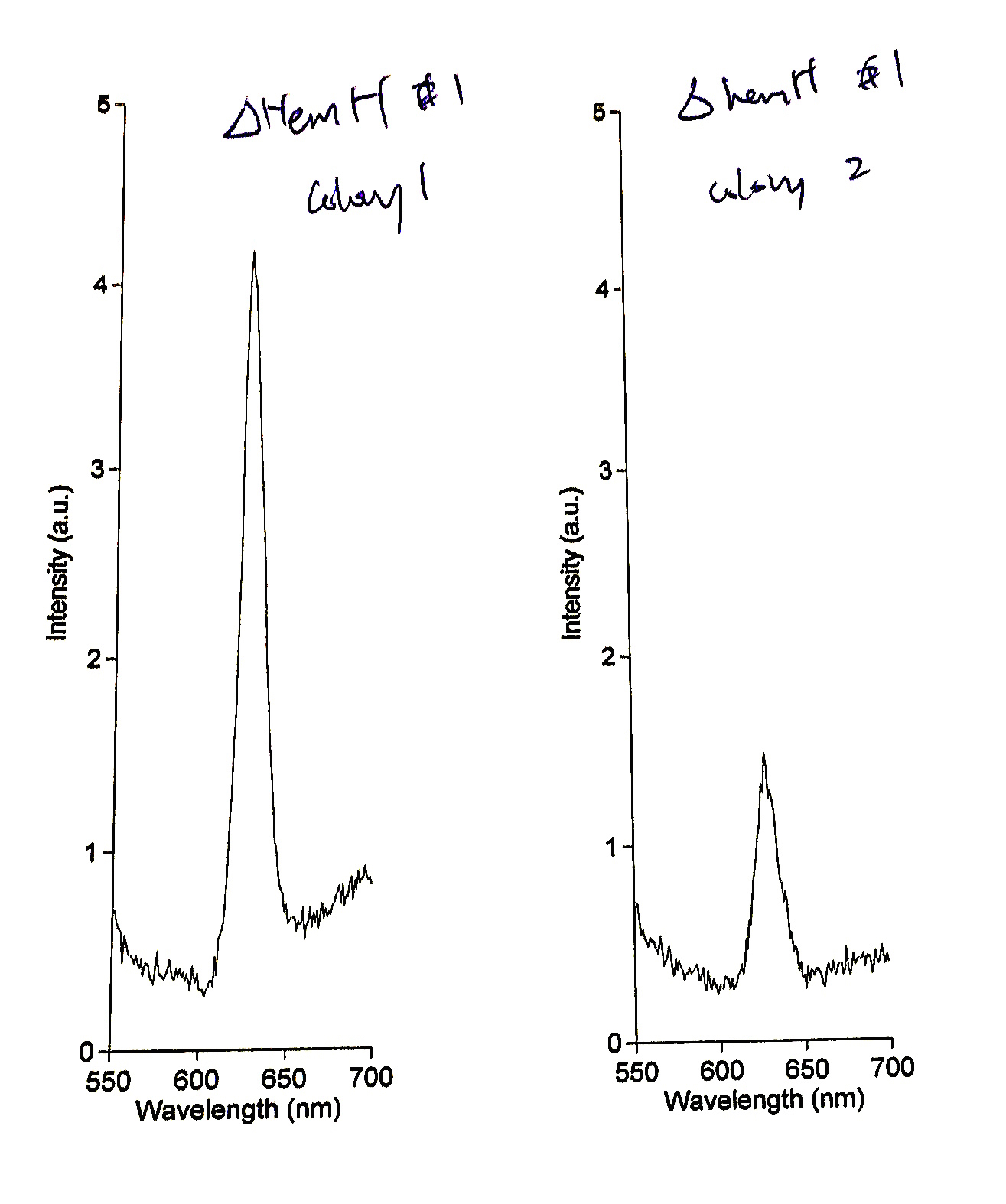
Fig 4. Mutant #1 was selected and further screened. Accumulation of PPIX was confirmed using further fluorescence spectroscopy assays.





