BBa_K2077002: pSB416 GPD mls-yeGFP
Overview
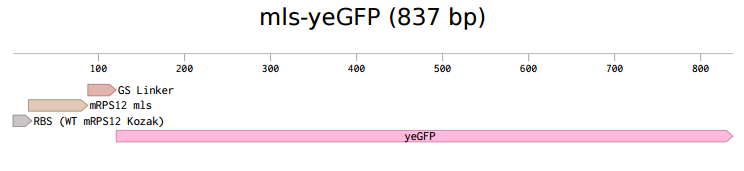
According to 2015 Rose-Hulman iGEM team, the coding sequence of mitochondrial ribosomal protein mRPS12 starts with a mitochondrial localization signal (mls). From previous research, it is possible that this mls protein is able to transport and direct any attached protein into the mitochondria. To verify the hypothesis brought up in 2015, we constructed a hybrid system with the mls protein and a reporter protein. Instead of using the regular GFP, yeast-enhanced green fluorescent protein (yeGFP) was used in this system as optimal gene expression can be achieved [1].
Methods & Design
The first step was to isolate mls fragment from pSBIC3 mRPS12 mls submitted last year. To do that, we digested the pSBIC3 mRPS12 mls plasmid with EarI and SpeI. After extracting DNA from the gel, we performed NEBuilder to insert yeGFP into the pSBIC3 vector, followed by transformation on LB-chloro plates. Then, we picked colonies from transformation plates and inoculated in LB-chloro broth overnight, followed by minipreping them. The diagnostic gel showed positive results on these minipreps, indicating that mls-yeGFP construct was successfully made. Next, we inserted the mRPS12 mls-yeGFP fragment into yeast vector pSB416 GPD vector and transformed the new plasmid into competent E. Coli cells. After picking colonies from LB-amp plate and inoculating them in LB-amp broth, we minipreped the culture and performed a diagnostic gel. The results of the diagnostic gel confirmed that the transformation of mls-yeGFP and pSB416 GPD was successful. Then we followed yeast transformation protocol to clone the new plasmid, pSB416 GPD mls-yeGFP, into S. cerevisiae cells and plate them on CSM-Ura for future fluorescence quantitative analysis. To prepare cells for microscopy, GFP fixation protocol was conducted. On the slide we prepared, the negative control was wild type yeast cells transformed with empty pSB416 GPD plasmid, while the test was yeast cells transformed with pSB416 GPD containing mls-yeGFP gene in it. With the help of Purdue University, a Nikon microscope was used to observe fluorescence in yeast mitochondria with an excitation wavelength of 488 nm and an emission wavelength of 525 nm. After localized florescence was observed we then used MIto ID Red florescent stain to verify that the florescence of mls-yeGFP is targeted to mitochondria.
RFC 10 Yeast Expression Vectors
BBa_K2077000: pSB416-GPD:RFC 10 Compatibile URA3-selectable S. cerevisiae Expression Vector Overview
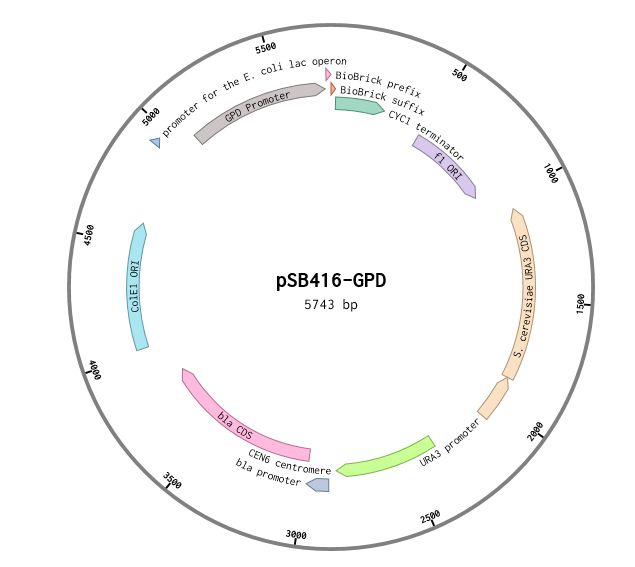
This is a RFC10 standard yeast expression vector adapted from the widely used plasmid p416-GPD. The MCS of p416-GPD was replaced with the standard BioBrick prefix and suffix and an illegal PstI restriction site near the URA3 promoter of p416-GPD was removed. In the resultant plasmid, pSB416-GPD (5743 bp), the BioBrick prefix and suffix is situated between the yeast GPD promoter and CYC1 terminator allowing for facile cloning and expression of translational units in yeast. M13 primers flank the transcriptional unit. The plasmid has a ColE1 origin and ampicillin resistance for replication in E.coli. It also contains a yeast CEN/ARS providing high fidelity maintenance and low copy number in yeast. We used this vector to express mRPS12 TU and mls-yeGFP in BY4741 S. cerevisiae.
Design
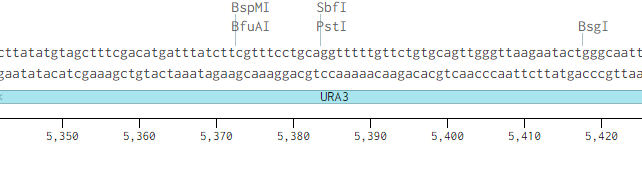
Originally p416-GPD had an illegal PstI site in the URA3 promoter region.
Removing the PstI site from p416 GPD required simply cutting with SbfI, blunting the sticky ends of the DNA removing the PstI site, and then ligating the plasmid back together. Giving us the resulting region with the PstI site removed.
We also needed to replace the MCS in pSB416-GPD with the RFC 10 standard prefix and suffix. We removed the MCS in p416-GPD by cutting the XbaI and XhoI and running a NEBuilder assembly with a fragment containing the BB prefix and suffix flanked with 40 base pairs of homology to the vector on both sides.
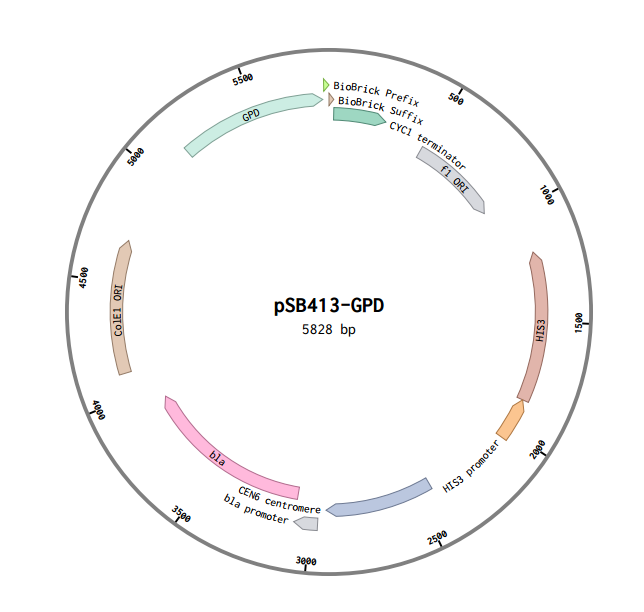
BBa_K2077001: pSB413-GPD: RFC 10 Compatible HIS3-selectable S. cerevisiae Expression Vector
Overview
This is a RFC10 standard yeast expression vector adapted from the widely used plasmid pSB413-GPD. The MCS of p413-GPD was replaced with the standard BioBrick prefix and suffix and an illegal PstI restriction site near the HIS3 promoter of p413-GPD was removed. In the resultant plasmid, pSB413-GPD (5828 bp), the BioBrick prefix and suffix is situated between the strong constitutive yeast GPD promoter (TDH3) and CYC1 terminator allowing for facile cloning and expression of translational units in yeast. Standard VF2 and VR primer binding sites are included and M13 primer binding sites bracket the promoter and terminator. The plasmid has a ColE1 origin and ampicillin resistance for replication in E. coli. It also contains a yeast CEN/ARS and HIS3-selectable marker to provide high fidelity maintenance and low copy number in S. cerevisiae.
Design
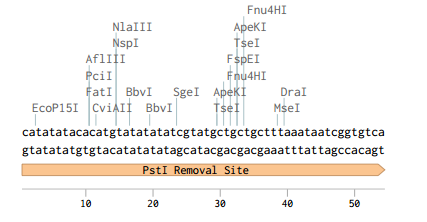
Just like in p416-GPD p413-GPD has an illegal PstI restriction site just outside of its selectable marker, HIS3. Unlike p416-GPD, however, this vector does not have and sbfI site surrounding its PstI site.
To remove the PstI site we decide to cut out a large chunk of the vector using its NheI and NsiI restriction sites and designed a replacement fragement with 30 base pairs of homology to the rest of the vector on both ends. We used a single nucleatide substitution to remove the PstI site from the replacement fragment. After following the NEBuilder protocol we successfully removed the PstI site resulting in the following sequence.
'We also needed to replace the MCS in pSB413-GPD with the RFC 10 standard prefix and suffix. We removed the MCS in p413-GPD by cutting the XbaI and XhoI and running a NEBuilder assembly with a fragment containing the BB prefix and suffix flanked with 40 base pairs of homology to the vector on both sides.