Testing mRPS12 TU
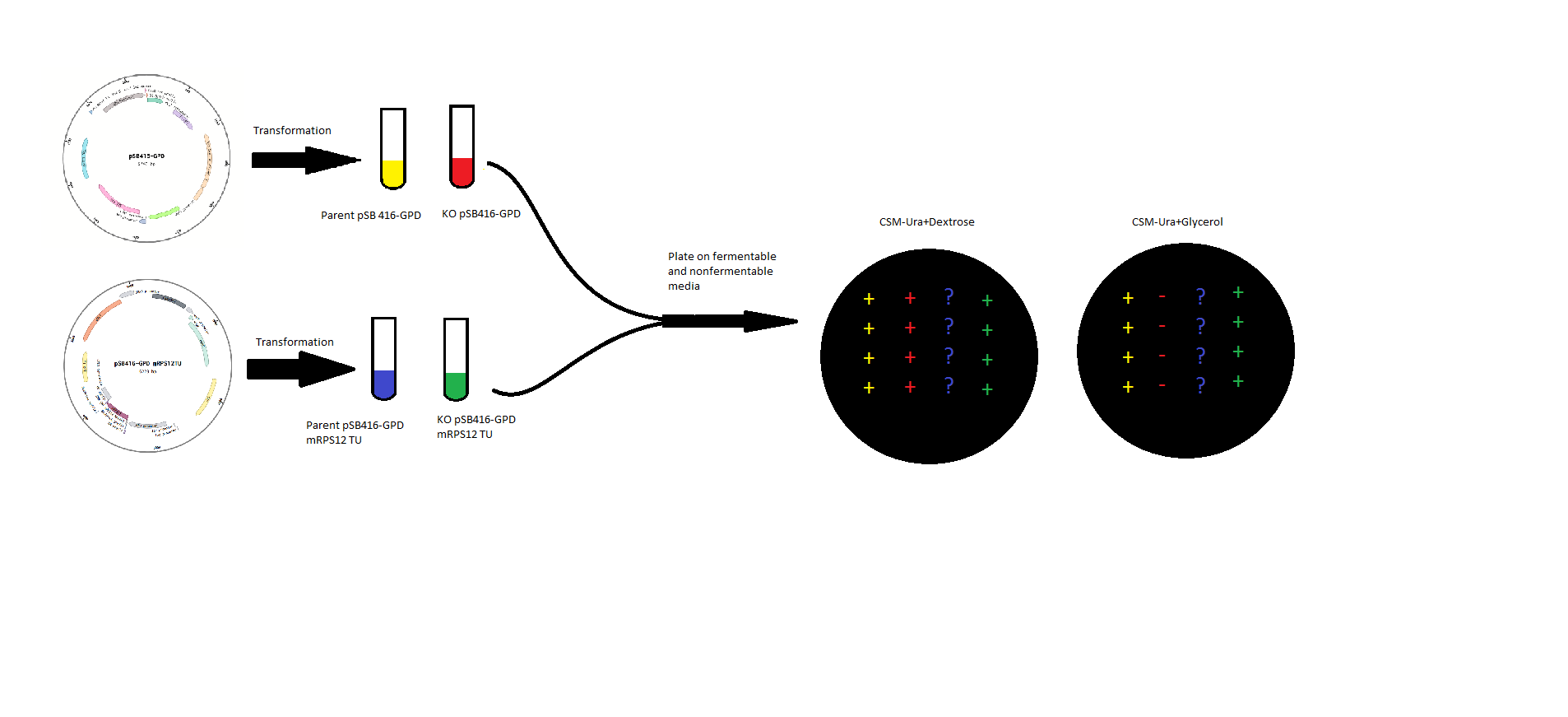
mRPS12 TU: mRPS12 TU was cloned into pSB416-GPD for expression in BY4741 Yeast and BY4741 mRPS12 TU knock out yeast. Competent yeast strains with or without mRPS12 were transformed with either pSB416-GPD with mRPS12 TU or empty pSB416-GPD and plated on CSM-Ura Plates. Colonies were allowed to grow for three days before they were picked and spotted onto two plates, one CSM-Ura with a fermentable Dextrose carbon source, and the other CSM-Ura but with a non-fermentable Glycerol carbon source. An experimental scheme is shown below.
Testing mRPS12 TU
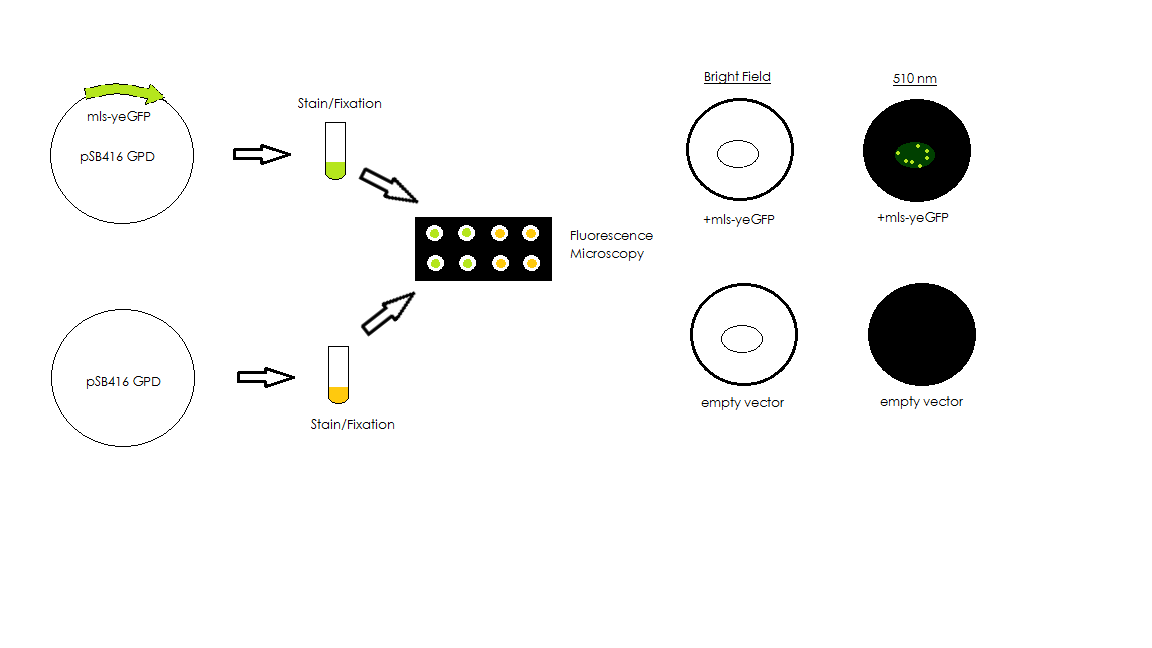
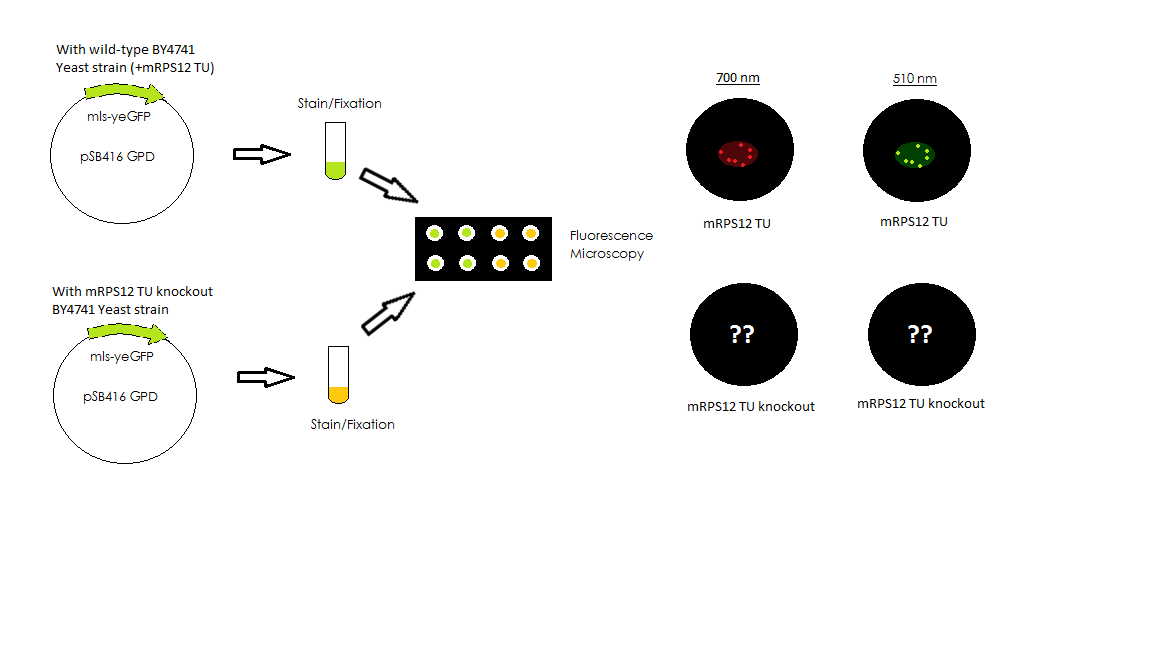
mls-yeGFP was cloned into pSB416-GPD for expression in BY4741 Yeast. Competent yeast cells were then transformed with either mls-yeGFP or empty pSB416 GPD vector and plated on CSM-Ura media to select for the presence of the pSB416-GPD plasmid. After three days of growth, colonies were picked and grown up in CSM-Ura liquid media for one day. Cultures were then fixed with paraformaldyhede for fluorescence microscopy. Using a fluorescence microscope, we compared brightfield and fluorescent views to determine if mls-yeGFP was causing transformed yeast to fluoresce. We then used a mitochondrial stain, Mito ID Red, to stain mitochondria in the wild-type BY4741 cells and compared brightfield and fluorescent views to determine if the mls actually targeted the GFP protein to the mitochondria in the yeast cells. Additionally, we used the Mito ID Red to stain BY4741 strains with mRPS12 TU gene knockout. The results from the knockout strain gave us insights on whether mitochondria were maintained, and if so, what would the function of mls change in response to that. The two figures below show the corresponding experimental scheme.