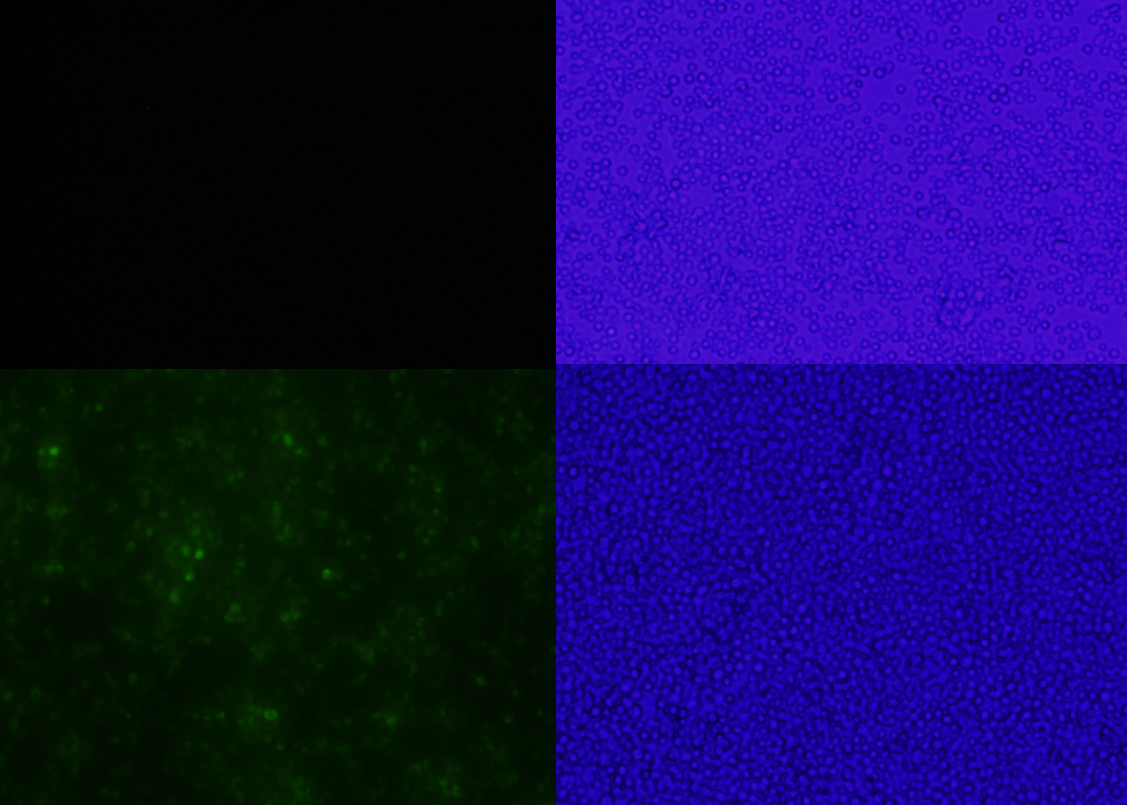
Figure 1: A comparison between yeast transformed with or without a florescent protein in pSB416-GPD Top left, BY4741 Yeast with empty pSB416-GPD exposed to 480nm light to test for florescence under 20x magnification. Top left, florescent view of the yeast without mls-yeGFP, top right the bright field view of the same strain. Bottom left, florescent view of yeast with mls-yeGFP in pSB416, bottom right the bright field view of the same strain.
Figure 2: Comparison between yeast with mRPS12, yeast with the gene knocked out, yeast with two copies of the gene, on in their genome and one in pSB416-GPD, and knock out yeast transformed with mRPS12 TU in our pSB416 GPD vector from right to left on each plate. This Picture was taken after two days of incubation.
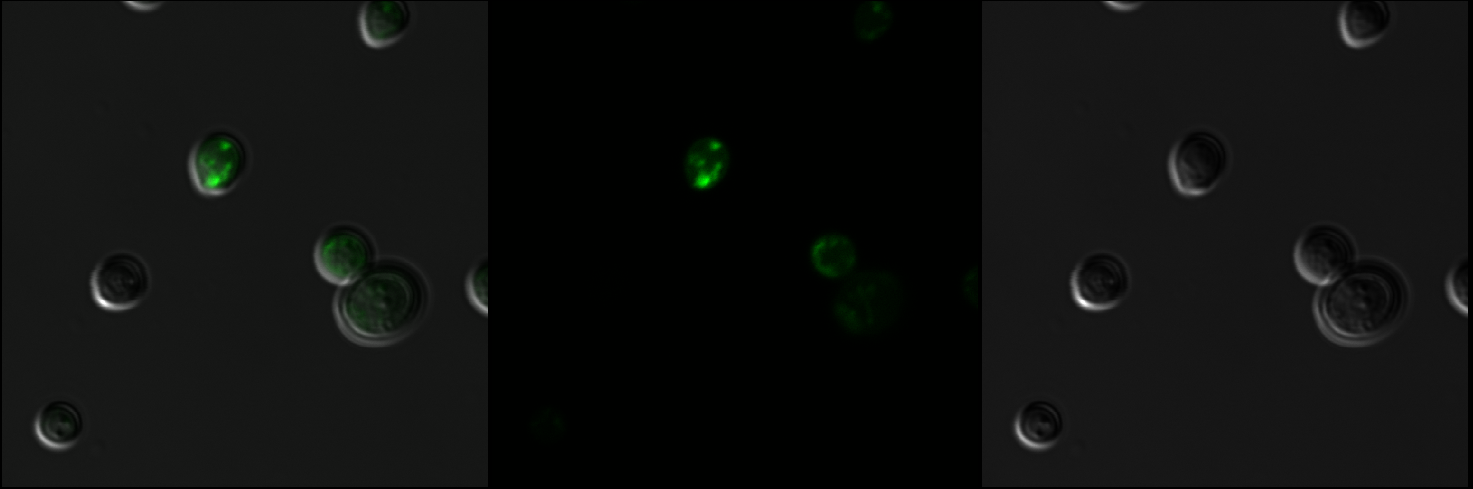
Figure 3: Observation of S. cerevisiae yeast cells that contain mls-yeGFP construct (test) with a 60X oil immersion objective. The right picture shows yeast cells under bright-field microscopy. The middle picture shows yeast cells under fluorescence microscopy. The left picture shows the overlay of the above mentioned two pictures.
Figure 4: Observation of S. cerevisiae yeast cells containing empty pSB416 GPD plasmid (negative control) with a 60X oil immersion objective. The right picture shows yeast cells under bright-field microscopy. The middle picture shows yeast cells under fluorescence microscopy. The left picture shows the overlay of the above mentioned two pictures.
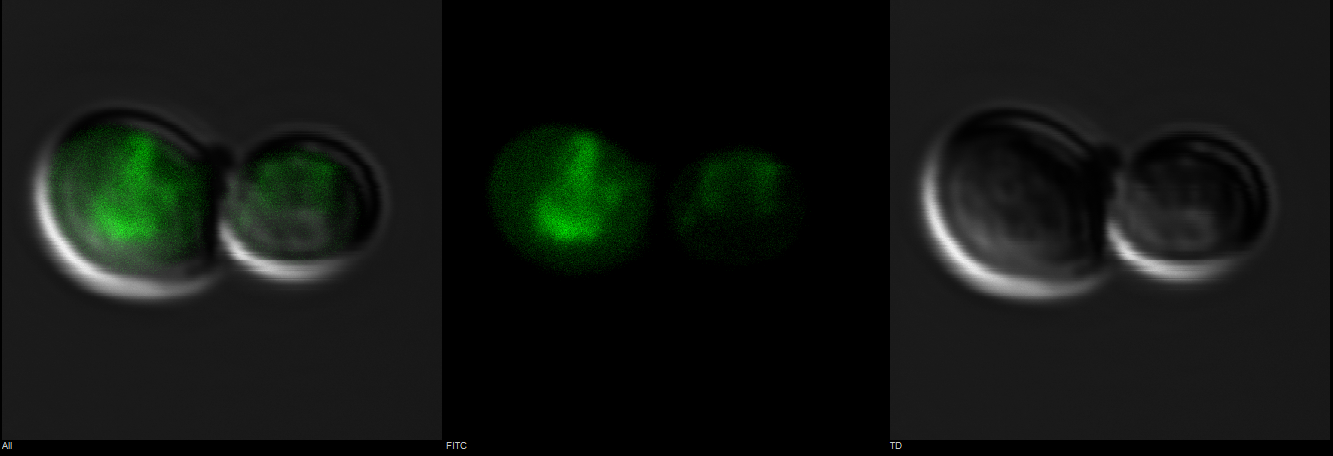
Figure 5: Observation of S. cerevisiae yeast cells that contain mls-yeGFP construct (test) with a 60X oil immersion objective. The right picture shows yeast cells under bright-field microscopy. The middle picture shows yeast cells under fluorescence microscopy. The left picture shows the overlay of the above mentioned two pictures.
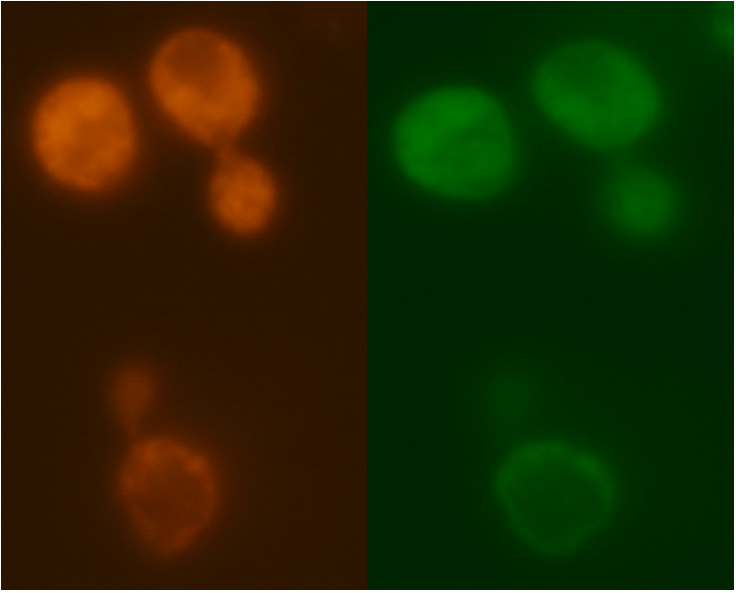
Figure 6: Comparison between Mito ID Red florescence, left, and mls-yeGFP florescence, right, in the mRPS12 TU knock out yeast strain.
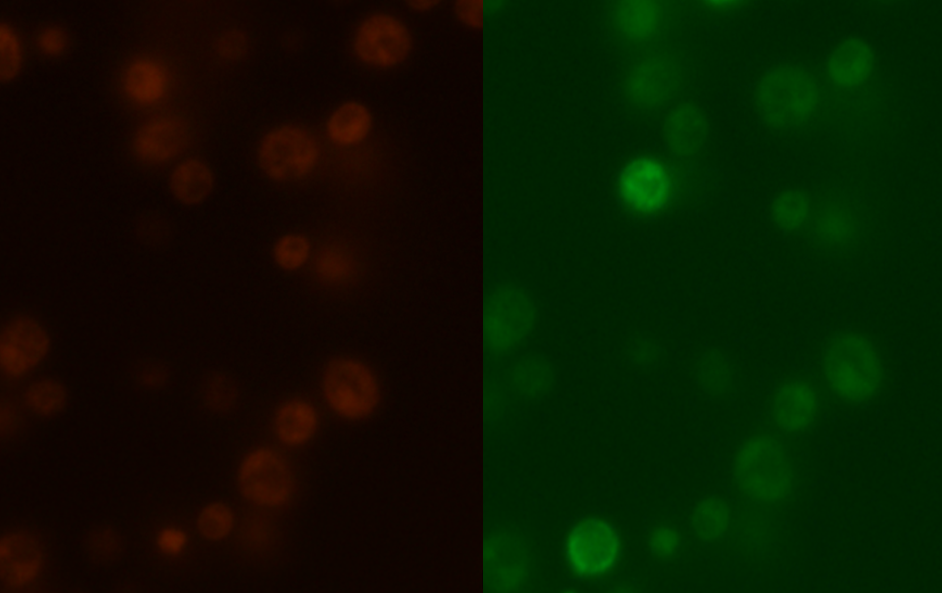
Figure 7: Comparison between Mito ID Red florescence, left, and mls-yeGFP florescence, right, in the mRPS12 TU wild type yeast strain.
The promoter, terminator, biobrick prefix and suffix, and selectable markers in pSB416-GPD all function as expected. Post cloning mls-yeGFP into the vector, E.coli transformed with it grew on LB+Amp plates verifying the function of the plasmids amp resistance. Yeast transformed with the vector grew in CSM-Ura media regardless of mls-yeGFP presence or absence in the vector, and yeast with mls-yeGFP fluoresce when excited with blue light, indicating that the Ura3 selectable marker, the GPD promoter, they CyC1 terminator, and the biobrick prefix and suffix function (Figure 1).
With all parts of the pSB416-GPD vector verified we proceeded to use it to test the complementation of mRPS12 TU. The gene failed to compliment in the knock out strain and restore the strains ability to grow on glycerol (Figure 2). No distinguishable difference in growth was present between the WT strain and the WT with empty pSB416 strain with an additional copy of the gene in pSB416-GPD. Overexpression of mRPS12 TU does have not have a great enough impact on mitochondrial function to cause yeast to become unable to grow on glycerol. The lack of impact the presence or absence of mRPS12 TU had on strain growth may indicate something wrong with the vector used, but no problems with gene expression were observed in the previous mls-yeGFP experiments that verified the function of the plasmid and the sequence of the vector with mRPS12 TU came back clean. The failure of complementation does not result from the loss of mitochondria in the knock out strain or the knock out strains mitochondria failing to import mRPS12 TU's preprotein mls signal (Figure 6). While without mRPS12 TU yeas cannot grow on non-fermentable carbon sources restoring the gene on a separate plasmid has failed to restore mitochondrial function in mRPS12 knock out yeast.
As shown in Figure 3, scattered green fluorescence was observed inside some yeast cells. According to the pattern of fluorescence, we are able to conclude that GFP is targeted to some internal organelles rather than cytoplasm, and it is possible that the fluorescence comes from mitochondria. However, additional experiments with mitochondria fluorescence dye is needed to show the actual location of mitochondria in order to fully prove the function of mls. Comparing to Figure 3, the negative control (Figure 4) does not emit any fluorescence under the same excitation condition, indicating that empty pSB416 GPD plasmid did not affect mls-yeGFP expression.A blown-up image of fluorescing yeast cells is shown below in Figure 5. It clearly shows that the intensity of fluorescence varies throughout the cells, indicating mls protein is targeting GFP to certain regions of the cell, which possibly would be the mitochondria. The targeted fluorescence pattern of mls-yeGFP matches the fluorescence pattern of mitochondria targeting red fluorescent stain Mito ID Red, verifying that the mRPS12 mls preprotein peptide does target proteins to mitochondria (Figures 6 & 7). Additionally mRPS12 mls targets proteins to the mitochondria regardless of whether or not a yeast strain can translate its mitochondrial genome and/or use its electron transport chain for respiration, as illustrated by the lack of difference between fluorescence pattern similarities between the red stain and mls-yeGFP in both the knock out strain and the parent strain (Figures 6 & 7).