| Line 42: | Line 42: | ||
<p><b>Testing mRPS12 TU</b></p> | <p><b>Testing mRPS12 TU</b></p> | ||
| + | |||
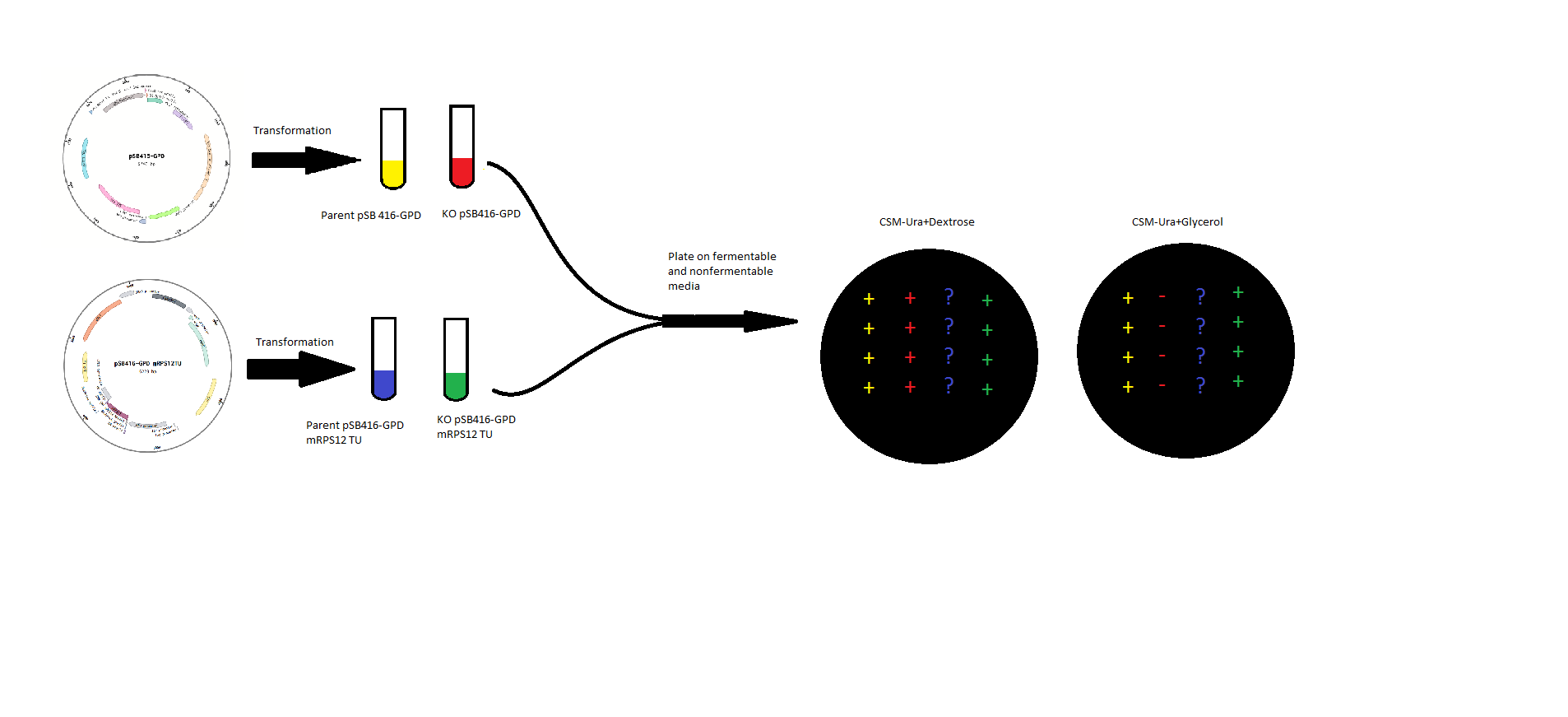
| + | <p>mRPS12 TU: mRPS12 TU was cloned into pSB416-GPD for expression in BY4741 Yeast and BY4741 mRPS12 TU knock out yeast. Competent mRPS12 + and – yeast strains were transformed with either pSB416-GPD with mRPS12 TU or empty pSB416-GPD and plated on CSM-Ura Plates. Colonies were allowed to grow for three days before they were picked and spotted onto two plates, one CSM-Ura with a fermentable Dextrose carbon source, and the other CSM-Ura but with a non-fermentable Glycerol carbon source</p> | ||
| + | |||
[[File:T--RHIT--MSPaintFigureOne.png]] | [[File:T--RHIT--MSPaintFigureOne.png]] | ||
| − | <p>mRPS12 TU | + | <p><b>Testing mRPS12 TU</b></p> |
| + | |||
| + | <p>mls-yeGFP was cloned into pSB416-GPD for expression in BY4741 Yeast. Competent yeast cells were then transformed with either mls-yeGFP or empty pSB416 GPD and plated on CSM-Ura media to select for the presence of the pSB416-GPD plasmid. After three days of growth, colonies were picked and grown up in CSM-Ura liquid media for one day. Cultures were then fixed with paraformaldyhede for fluorescence microscopy. Using a fluorescence microscope, we compared brightfield and fluorescent views to determine if mls-yeGFP was causing transformed yeast to fluoresce. We then used a mitochondrial stain, Janus Green, to stain mitochondria in the mls-yeGFP cells and compared brightfield and fluorescent views to determine if the mls actually targeted the GFP protein to the mitochondria in the yeast cells.</p> | ||
Revision as of 19:46, 1 October 2016
Describe the experiments, research and protocols you used in your iGEM project.
What should this page contain?
- Protocols
- Experiments
- Documentation of the development of your project
Inspiration
Testing mRPS12 TU
mRPS12 TU: mRPS12 TU was cloned into pSB416-GPD for expression in BY4741 Yeast and BY4741 mRPS12 TU knock out yeast. Competent mRPS12 + and – yeast strains were transformed with either pSB416-GPD with mRPS12 TU or empty pSB416-GPD and plated on CSM-Ura Plates. Colonies were allowed to grow for three days before they were picked and spotted onto two plates, one CSM-Ura with a fermentable Dextrose carbon source, and the other CSM-Ura but with a non-fermentable Glycerol carbon source
Testing mRPS12 TU
mls-yeGFP was cloned into pSB416-GPD for expression in BY4741 Yeast. Competent yeast cells were then transformed with either mls-yeGFP or empty pSB416 GPD and plated on CSM-Ura media to select for the presence of the pSB416-GPD plasmid. After three days of growth, colonies were picked and grown up in CSM-Ura liquid media for one day. Cultures were then fixed with paraformaldyhede for fluorescence microscopy. Using a fluorescence microscope, we compared brightfield and fluorescent views to determine if mls-yeGFP was causing transformed yeast to fluoresce. We then used a mitochondrial stain, Janus Green, to stain mitochondria in the mls-yeGFP cells and compared brightfield and fluorescent views to determine if the mls actually targeted the GFP protein to the mitochondria in the yeast cells.