m |
m |
||
| Line 43: | Line 43: | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
<div class = "page_references"> | <div class = "page_references"> | ||
| − | <p> References</p> | + | <p>References</p> |
<ol> | <ol> | ||
<li>Lin, Steven, and John E. Cronan. "Closing in on complete pathways of biotin biosynthesis." Molecular BioSystems 7.6 (2011): 1811-1821. </li> | <li>Lin, Steven, and John E. Cronan. "Closing in on complete pathways of biotin biosynthesis." Molecular BioSystems 7.6 (2011): 1811-1821. </li> | ||
</ol> | </ol> | ||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
</div> | </div> | ||
Revision as of 19:54, 18 October 2016
Proof of Concept
Measuring biotin production to calculate reduced electron donors
After identifying the genes and constructing the electron donor overproducing plasmids, proper assays were necessary in order to see if our new cells produced usable forms of both ATP and electron donors. Separate assays were used to test this concept. Not only was it shown that the products were overproduced, but also significant data supported the claim that these products were usable in protein synthesis.
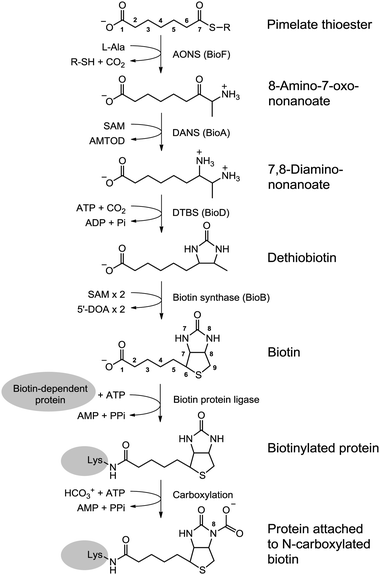
After analyzing the results of our absorbance data of electron donors, it was decided to go a step further to ensure that these construct produced usable electron donors. A biotin assay was found that offered an indirect way of measuring the amount of reduced electron donors present. Biotin, commonly know as vitamin B7, is produced by a pathway that uses reduced flavodoxin/ferredoxin,specifically the SAM pathway that turns dethiobiotin to biotin[1] (see diagram below).
By measuring the amount of biotin in the new cells, and recognizing the 2:1 stoichiometric relationship between reduced electron donors and biotin molecules, the amount of electron donors present and used can be indirectly measured. Results showed that overall, our constructs worked to increase biotin production, as seen in the graphs below.
- When our constructs were in DH10B cells, the fldA, fldA/pfo, petF, and petF/pfo strains produced more biotin than the control DH10B strain at some induction level
- When our constructs were in the ΔaceE knockout cells, the fldA, fldA/pfo, and petF strains all produced more biotin than the control DH10B strain, control ΔaceE knockout strain, and DH10B+constructs strain (petF/pfo data is lacking due to its inability to grow)


Go to the conclusions page to see a complete summary of our results and proof of concept experiments.
References
- Lin, Steven, and John E. Cronan. "Closing in on complete pathways of biotin biosynthesis." Molecular BioSystems 7.6 (2011): 1811-1821.





