(Prototype team page) |
|||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{RHIT}} | {{RHIT}} | ||
| − | |||
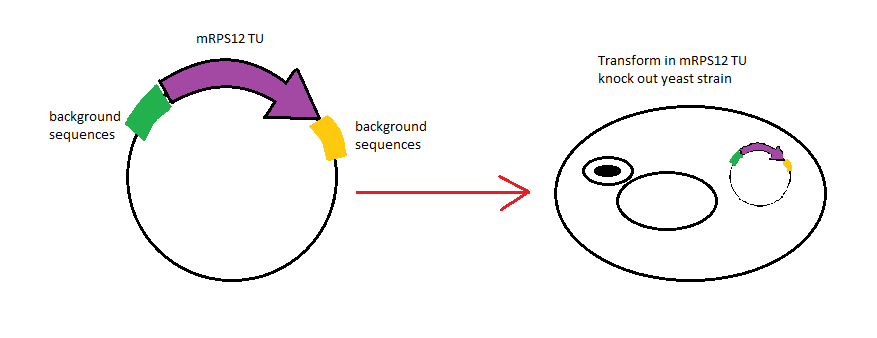
| + | <p> In this project, we successfully produced two yeast expression vectors with BioBrick prefix and suffix. Using the vectors we produced, we managed to verify function of mitochondrial localization signal (mls) by testing the hybrid construct mls-yeGFP under a fluorescence microscope. Expression of mls-yeGFP causes mitochondria targeted fluorescence regardless of whether the strain has functional mitochondrial ribosomes and a functional electron transport chain. Unfortunately, experimental results indicated that expressing the mitoribosomal protein mRPS12 in a haploid mRPS12 knockout strain did not restore aerobic respiration and their ability to grow on non-fermentable carbon sources. We replicated this experiment several times while the same results were obtained. Expressing mRPS12 in wild type S. <i>cerevisiae</i> did not result any adverse effect, therefore we can conclude that its failure to restore mitochondria function in the knock out strain did not result from overexpression of the gene behind the GPD promoter. Therefore we hypothesized that the knock out strain we used has lost its mitochondrial genome which explains why restoring mitoribosomal function does not restore the strains ability to process glycerol. We continued our investigation using mitochondria targeting red fluorescent stain, Mito ID Red. Microscopy illustrated that the mRPS12 TU knock out strain still had mitochondria, and that the preprotein mitochondria targeting peptide from mls-yeGFP still function in that strain. We purpose that the failure of mRPS12 TU complementation results from the loss of background sequence before and after the TU that could impact properties of the mRPS12 protein such as its half life. To further investigate this we will clone mRPS12 TU and its surrounding genomic DNA into a vector and try to use it to complement the mRPS12 TU knock out phenotype.</p> | ||
| − | + | [[File:T--RHIT--demonstrate.png]] | |
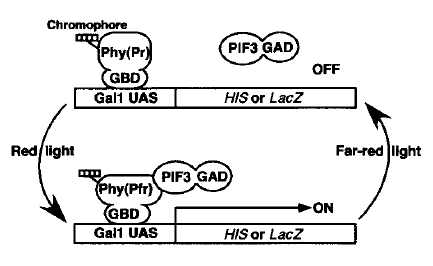
| − | <p> | + | <p>Once the function of mRPS12 is verified, we would like to connect a light-switchable system [1] to the mRPS12 gene in order to achieve remote control. Essentially the system utilizes protein-protein interaction between phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3) to control gene expression. By attaching a chromophore phycocyanobilin (PCB) extracted from cyanobacterium to PhyB, exposure to red light would change conformation of PhyB from ground state Pr to active state Pfr. Such conformation change would alter interaction between PhyB and PIF3, resulting in turning the following gene on or off. This system is originally proposed in 2002 and it was based on Gal4 promoter [1], while an iGEM team from Munich modified it by replacing Gal4 promoter with LexA [2]. The advantage of using LexA is that, since LexA comes from prokaryotes, it minimizes the interference between endogenous yeast metabolism [2]. |
| + | </p> | ||
| + | [[File:T--RHIT--Chrmophore.png]] | ||
| − | < | + | <p>References:</p> |
| − | + | <p>[1] S. Shimizu-Sato, E. Huq, J. Tepperman and P. Quail, "A light-switchable gene promoter system", Nature Biotechnology, vol. 20, no. 10, pp. 1041-1044, 2002.</p> | |
| − | + | <p>[2] http://parts.igem.org/Part:BBa_K801043</p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 03:38, 20 October 2016
In this project, we successfully produced two yeast expression vectors with BioBrick prefix and suffix. Using the vectors we produced, we managed to verify function of mitochondrial localization signal (mls) by testing the hybrid construct mls-yeGFP under a fluorescence microscope. Expression of mls-yeGFP causes mitochondria targeted fluorescence regardless of whether the strain has functional mitochondrial ribosomes and a functional electron transport chain. Unfortunately, experimental results indicated that expressing the mitoribosomal protein mRPS12 in a haploid mRPS12 knockout strain did not restore aerobic respiration and their ability to grow on non-fermentable carbon sources. We replicated this experiment several times while the same results were obtained. Expressing mRPS12 in wild type S. cerevisiae did not result any adverse effect, therefore we can conclude that its failure to restore mitochondria function in the knock out strain did not result from overexpression of the gene behind the GPD promoter. Therefore we hypothesized that the knock out strain we used has lost its mitochondrial genome which explains why restoring mitoribosomal function does not restore the strains ability to process glycerol. We continued our investigation using mitochondria targeting red fluorescent stain, Mito ID Red. Microscopy illustrated that the mRPS12 TU knock out strain still had mitochondria, and that the preprotein mitochondria targeting peptide from mls-yeGFP still function in that strain. We purpose that the failure of mRPS12 TU complementation results from the loss of background sequence before and after the TU that could impact properties of the mRPS12 protein such as its half life. To further investigate this we will clone mRPS12 TU and its surrounding genomic DNA into a vector and try to use it to complement the mRPS12 TU knock out phenotype.
Once the function of mRPS12 is verified, we would like to connect a light-switchable system [1] to the mRPS12 gene in order to achieve remote control. Essentially the system utilizes protein-protein interaction between phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3) to control gene expression. By attaching a chromophore phycocyanobilin (PCB) extracted from cyanobacterium to PhyB, exposure to red light would change conformation of PhyB from ground state Pr to active state Pfr. Such conformation change would alter interaction between PhyB and PIF3, resulting in turning the following gene on or off. This system is originally proposed in 2002 and it was based on Gal4 promoter [1], while an iGEM team from Munich modified it by replacing Gal4 promoter with LexA [2]. The advantage of using LexA is that, since LexA comes from prokaryotes, it minimizes the interference between endogenous yeast metabolism [2].
References:
[1] S. Shimizu-Sato, E. Huq, J. Tepperman and P. Quail, "A light-switchable gene promoter system", Nature Biotechnology, vol. 20, no. 10, pp. 1041-1044, 2002.
[2] http://parts.igem.org/Part:BBa_K801043