XintongXia (Talk | contribs) |
XintongXia (Talk | contribs) |
||
| Line 14: | Line 14: | ||
<p><b>Method</b></p> | <p><b>Method</b></p> | ||
<p> | <p> | ||
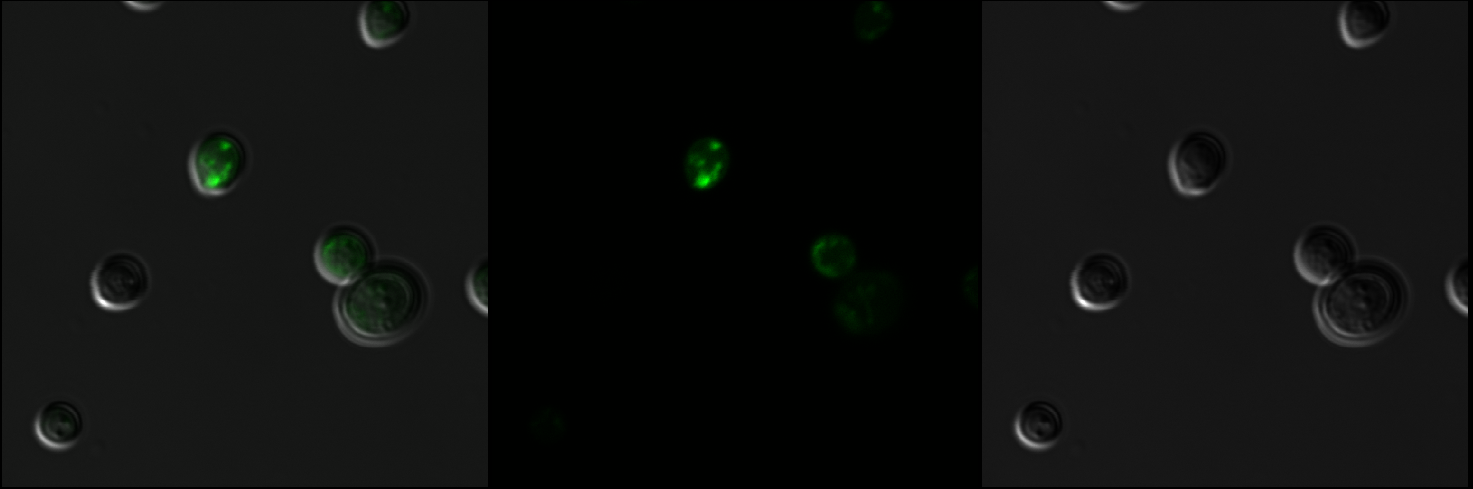
| − | The first step was to isolate mls fragment from pSBIC3 mRPS12 mls submitted last year. To do that, we digested the pSBIC3 mRPS12 mls plasmid with EarI and SpeI. After extracting DNA from the gel, we performed NEBuilder to insert yeGFP into the pSBIC3 vector, followed by transformation on LB-chloro plates. Then, we picked colonies from transformation plates and inoculated in LB-chloro broth overnight, followed by minipreping them. The diagnostic gel showed positive results on these minipreps, indicating that mls-yeGFP construct was successfully made. Next, we inserted the mRPS12 mls-yeGFP fragment into yeast vector pSB416 GPD vector and transformed the new plasmid into competent E. Coli cells. After picking colonies from LB-amp plate and inoculating them in LB-amp broth, we minipreped the culture and performed a diagnostic gel. The results of the diagnostic gel confirmed that the transformation of mls-yeGFP and pSB416 GPD was successful. Then we followed yeast transformation protocol to clone the new plasmid, pSB416 GPD mls-yeGFP, into S. cerevisiae cells and plate them on CSM-Ura for future fluorescence quantitative analysis. An Olympus 1X51 microscope was used to observe fluorescence in yeast | + | The first step was to isolate mls fragment from pSBIC3 mRPS12 mls submitted last year. To do that, we digested the pSBIC3 mRPS12 mls plasmid with EarI and SpeI. After extracting DNA from the gel, we performed NEBuilder to insert yeGFP into the pSBIC3 vector, followed by transformation on LB-chloro plates. Then, we picked colonies from transformation plates and inoculated in LB-chloro broth overnight, followed by minipreping them. The diagnostic gel showed positive results on these minipreps, indicating that mls-yeGFP construct was successfully made. Next, we inserted the mRPS12 mls-yeGFP fragment into yeast vector pSB416 GPD vector and transformed the new plasmid into competent E. Coli cells. After picking colonies from LB-amp plate and inoculating them in LB-amp broth, we minipreped the culture and performed a diagnostic gel. The results of the diagnostic gel confirmed that the transformation of mls-yeGFP and pSB416 GPD was successful. Then we followed yeast transformation protocol to clone the new plasmid, pSB416 GPD mls-yeGFP, into S. cerevisiae cells and plate them on CSM-Ura for future fluorescence quantitative analysis. An Olympus 1X51 microscope was used to observe fluorescence in yeast mitochondria.</p> |
<p><b>Results</b></p> | <p><b>Results</b></p> | ||
Revision as of 20:32, 10 August 2016
A basic part is a functional unit of DNA that cannot be subdivided into smaller component parts. <a href="http://parts.igem.org/wiki/index.php/Part:BBa_R0051">BBa_R0051</a> is an example of a basic part, a promoter regulated by lambda cl.
Most genetically-encoded functions have not yet been converted to BioBrick parts. Thus, there are many opportunities to find new, cool, and important genetically encoded functions, and refine and convert the DNA encoding these functions into BioBrick standard biological parts.
--------------------------------------------------------------------------------------------------------------------------------------------------------------------
pSB416 GPD mls-yeGFP
Overview
According to 2015 Rose-Hulman iGEM team, the coding sequence of mitochondrial ribosomal protein mRPS12 starts with a mitochondrial localization signal (mls). From previous research, it is possible that this mls protein is able to transport and direct any attached protein into the mitochondria. To verify the hypothesis brought up in 2015, we constructed a hybrid system with the mls protein and a reporter protein. Instead of using the regular GFP, yeast-enhanced green fluorescent protein (yeGFP) was used in this system as optimal gene expression can be achieved [1].
Method
The first step was to isolate mls fragment from pSBIC3 mRPS12 mls submitted last year. To do that, we digested the pSBIC3 mRPS12 mls plasmid with EarI and SpeI. After extracting DNA from the gel, we performed NEBuilder to insert yeGFP into the pSBIC3 vector, followed by transformation on LB-chloro plates. Then, we picked colonies from transformation plates and inoculated in LB-chloro broth overnight, followed by minipreping them. The diagnostic gel showed positive results on these minipreps, indicating that mls-yeGFP construct was successfully made. Next, we inserted the mRPS12 mls-yeGFP fragment into yeast vector pSB416 GPD vector and transformed the new plasmid into competent E. Coli cells. After picking colonies from LB-amp plate and inoculating them in LB-amp broth, we minipreped the culture and performed a diagnostic gel. The results of the diagnostic gel confirmed that the transformation of mls-yeGFP and pSB416 GPD was successful. Then we followed yeast transformation protocol to clone the new plasmid, pSB416 GPD mls-yeGFP, into S. cerevisiae cells and plate them on CSM-Ura for future fluorescence quantitative analysis. An Olympus 1X51 microscope was used to observe fluorescence in yeast mitochondria.
Results
References:
[1]B. Cormack, G. Bertram, M. Egerton, N. Gow, S. Falkow and A. Brown, "Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans", Microbiology, vol. 143, no. 2, pp. 303-311, 1997.