| Line 394: | Line 394: | ||
<h2>6/27/16</h2> | <h2>6/27/16</h2> | ||
| + | <ul>(XX) Goal: Redo digest and harvest TU and mls</ul> | ||
| + | <ul>Major steps:</ul> | ||
| + | <ul> | ||
| + | <ul>Cut mRPS12 TU with EcoRI/PstI</ul> | ||
| + | <ul>Cut Mrps12 mls with EarI/SpeI</ul> | ||
| + | </ul> | ||
| + | <ul>Gel order: L,TU1a, TU1b, TU2a, TU2b, mls1a, mls1b, mls2a, mls2b</ul> | ||
| + | <ul>Gel results:</ul> | ||
| + | <ul> | ||
| + | <ul>harvested TU bands from TU2a and TU2b (2 bands per tube)</ul> | ||
| + | <ul> | ||
| + | <ul>TU gel extract = 0.363 g</ul> | ||
| + | </ul> | ||
| + | <ul>harvested four mls bands (2 bands per tube)</ul> | ||
| + | <ul> | ||
| + | <ul>mls gel extract A = 0.342 g</ul> | ||
| + | <ul>mls gel extract B = 0.255 g</ul> | ||
| + | </ul> | ||
| + | </ul> | ||
| + | </ul> | ||
| + | <ul>Other activities:</ul> | ||
| + | <ul> | ||
| + | <ul>QIA gel extraction kit</ul> | ||
| + | <ul>Used nanodrop to test DNA amount:</ul> | ||
| + | <ul> | ||
| + | <ul>TU = 3.1 ng/ | ||
| + | <ul>mls A = 41.2 ng/ | ||
| + | <ul>mls B = 27.7 ng/ | ||
| + | </ul> | ||
| + | <ul>*according to nanodrop readings, TU needs to be precipitate while we run NEBuilder on mls A and mls B</ul> | ||
| + | </ul> | ||
| + | (HC) Ran NEBuilder on P413 GPD and 16-1 fragment. | ||
| + | 1μL DNA | ||
| + | 3μL fragment | ||
| + | 10μL NEBuilder HiFi DNA Assembily Master Mix | ||
| + | 6μL Sterile DI water | ||
| + | |||
| + | |||
| + | (JM/CH) | ||
| + | |||
| + | The first thing we are doing is running the blunt end ligation protocol using NEB Quick Blunting Kit and the NEB Quick Ligation Kit | ||
| + | |||
| + | This was done with 3 tubes of 416 CYC1 and 3 tubes of 426 GPD | ||
| + | |||
| + | Next we did a transformation on one of the 416 CYC1 and one of the 426 GPD | ||
| + | |||
| + | Those are spread on plates and are now incubating so we can use them tomorrow | ||
Revision as of 18:28, 18 August 2016
NOTEBOOK.
I should probably put some Table of Contents up here, so one could easily skip from day to day... at any rate, here's the current info dump.
Heads-up: I know there aren't circles on the list. This is apparently harder to do than I had envisioned?
6/13/16
- (HC) Made LB-Amp plates
- 450ml of DI water
- 5.0g tryptone
- 2.5g yeast
- 5.0g NaCl
- 7.5g Agar
- This was then adjusted to a pH of 7.0 using NaOH. It was then autoclaved. 2.5mL of 10mg/mL of Amp for a final concentration of 50 μg/mL was achieved.
6/14/16
- (HC) made CSM -His plates
6/15/16
- (XX) Goal: Run a gel to check whether pSB416 GPD has PstI site removed (diagnostic gel)
- 1. pSB416 GPD cut wit AvaI
- 2. pSB416 GPD cut with PstI
- 3. p146GPD cut with AvaI
- 4. p416GPD cut with PstI
- Major steps:
- Digest:
6 sterile water, 1 CutSmart buffer, 2 DNA, 1 enzyme (AvaI or PstI-HF)
- Gel Order: L, 1, 3, 2, 4
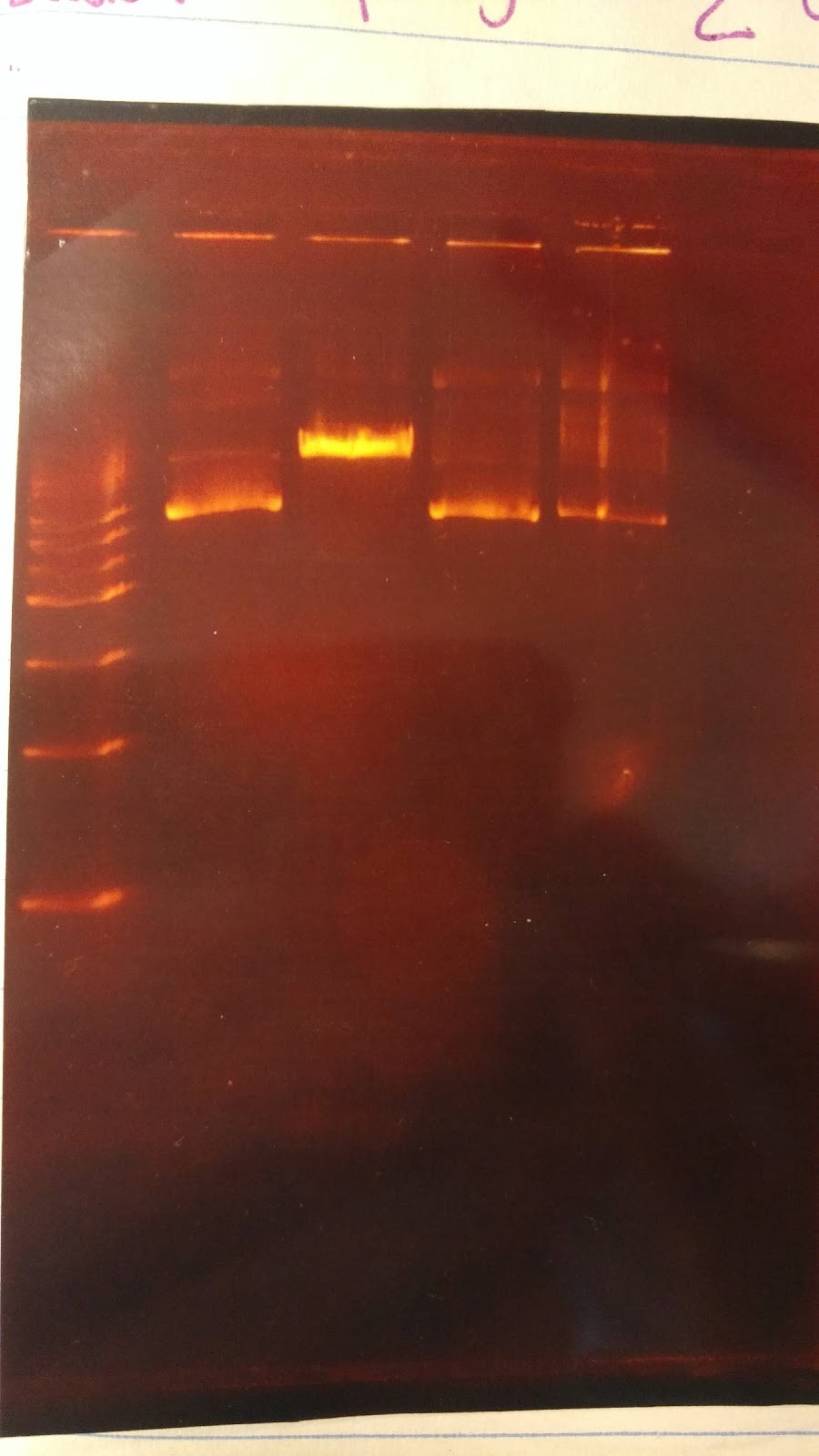
- Expected results on this diagnostic gel:
- 1. No cut because no AvaI site in pSB416 GPD
- 2. Should only see one cut because only one PstI site
- 3. Should see one cut
- 4. Should see two bands
- Experimental results:
Gel results are not good; need to rerun this diagnostic gel
- Other Activities:
- Created mRPS12+/KanMZ- (parent) and mRPS12-/KanMX+ (mutant) master plate on YPD
- Replica plating YPG (Should this be YPD???) , YPG, -His, -Ura, -Ura+Glycerol, YPD+Paro, YPG+Paro
6/16/16
(HC) Ran a boiling and spin prep on P413 GPD
- Boiling Prep Procedure:
- 1) Pellet cells from 1.5mL of an overnight L-Amp culture of transformed E. coli cells in a microcentrifuge for 20-30 seconds and remove supernatant.
- 2) Use a toothpick and vortexer to resuspend the cells in 100 μl(0.1mL) of STET buffer(8% sucrose; 5%Triton X-100;50 mM EDTA; Tris-Cl, pH 8.0) containing 1mg/mL lysozyme(prepare 1 mg/mLenzyme in STET just prior to use; 1mL= 10 preps).
- 3)Place the suspension in a boiling water bath for 90 seconds.
- 4) Spin in a microcentrifuge for 15 min at the highest speed.
- 5)Remove the pellet (a gelatinous mass of cell debris) with a toothpick and discard.
- 6) Precipitate nucleic acid at -20℃ for 30 minutes using an equal volume of isopropanol.
- 7) Spin the tube in a microcentrifuge for 10 minutes at RTo to pellet the nucleic acids.
- 8) Was pellet twice with 70% ethanol and dry thoroughly.
- 9) Resuspend the pellet in 50 μL of sterile DI water or TE buffer.
- Spin Prep: We used the QIAprep Spin Miniprep Kit to run a spin prep and followed the instructions inside.
- (XX)Goal: Rerun the diagnostic gel stated yesterday (6/15/16)
- Experimental results:
Good gel results, confirming that the PstI site has been removed in pSB416 GPD. pSB416 ready to be sequenced.
- Other activities:
Incubated pSBIC3 mRPS12 TU and pSBIC3 mRPS12 mls (from last year iGEM team??) in LB-chloramphenicol broth @37C overnight at 250 rpm
6/17/16
(HC) Ran two digests for P413 GPD
- Analytical Digest:
- 5 μL of DNA(Boiling Prep)
- 1μL of PstI HF
- 1μL of CutSmart buffer
- 3μL of sterile DI water
- Preparative Digest:
- 7μL of DNA(Spin Prep)
- 1μL of NheI
- 1μL of NsiI
- 1μL of CutSmart buffer
- Gel Results:
- PREP GEL
- Lane 1: Ladder
- Lane 3: Preparative Digest
- INSERT NOTEBOOK PICTURE #001 HERE
- Lane 1: Ladder
- Lane 2: Uncut (2μL of DNA and 8μL of water)
- Lane 3: Analytical Digest
- INSERT NOTEBOOK PICTURE #002 HERE
- (XX) Master plate results:
- Nothing grew on control plates (-His, -Ura, -Ura+Glycerol)
- Both parent and mutant grew on YPD and YPD+Paro, indicating that Paro has no effect on yeast growth
- Mutant cannot grow on YPG or YPG+Paro because of the mRPS12 gene knockout
- Other Activities:
- Cut p413GPD with NheI and NsiI to get rid of the illegal PstI site
- Massive gel to run boiling preps
- Conclusion:
- P413GPD cut looks like an uncut plasmid
- Massive gel has many blurry bands, which indicates that boiling preps are very impure
6/20/16
- (XX) Unltimate Goal:
- Put pSBIC3 mRPS12 TU into our own standardized yeast vectors
- Validate mls function by creating mls-yeGFP construct
- Goal for today:
- Digest and Harvest mRPS12 TU from pSBIC3(tube 1)
- 5μL of DNA
- 1μL of EcoRI-HF
- 1μL of pstI-HF
- 1μL of CutSmart buffer
- 2μL of sterile water
- Digest and Harvest mRPS12 mls from pSBIC3 (tube 2)
- 5μL of DNA
- 1μL of EarI
- 1μL of SpeI
- 1μL of CutSmart buffer
- 2μL of sterile water
- Gel Order: L, 1C, 1XX, 1XL, 2C, 2XX, 2XL
- **C-uncut plasmid, XX-Xintong, XL-Xander
- Gel results:
- Uncut does not show up on the gel
- Not a very good results but we harvested the bands anyway
- Other activiites:
- Excise bands from the gel and perform gel extraction (QIA kit):
- Tube 1 = 0.26 g
- Tube 2 = 0.14 g
- Tube 3 = 0.17 g
- *tube 3 contains p413 GPD with NsII/NheI cut
- Prepare Yeast Competent Cells
- 10 mL YPD, inoculated at 250RPM @ 30C
- Both BY4741 and BY4741 mRPS12 is prepared
6/21/16
- (XX) Goal for Today:
- DNA Assembly
- Major Steps:
- We used nanodrop to determine the amount of DNA in tube 1 (TU) and tube 2 (mls)
- Tube 1: 4.5 ng/
- Tube 2: 4.0 ng/
- * Nano drop results indicate that we need to precipitate DNA for further experiment
- After precipitation, nanodrop reports 12.6 ng/ DNA
- NEBuilder HiFi DNA Assembly Cloning Kit:
- 2μL Ear/SpeI digest pSBIC3 mls
- 8μL fragment 16-4 (yeGFP)
- 10μL HiFi Master
- *All three above, set the reaction on ice; 1:2 vecotr: insert
- Thermocycler
- Other Activities:
- Yeast competent cell from yesterday did not grow
- Prepare new yeast competent cells: 5 mL YPD in round bottom tubes
6/22/16
- (XX)Goal:
- Prepare Yeast Competent cells
- Diagnostic gel on mRPS12 TU minipreps
- Major Steps:
- Yeast competent cells:
- Spectromphotometer OD 260:
- 1000 YPD Blank
- 1000 KnockOut(KO) has 1.777 Abs.
- 1000 Parent has 2.222 Abs.
- Regrow these cells into 15 mL broth for about 2 hrs:
- Take 0.5 mL current culture into 14.5 mL YPD
- Spectrophotometer Results:
- KO: 0.390 Abs.
- Parent: 0.692 Abs.
- Follow yeast competent cell protocol
- Diagnostic Gel:
- 1: pSBIC3 mRPS12 TU miniprep 1 cut with EcoRI and PstI
- 2: pSBIC3 mRPS12 TU miniprep 2 cut with EcoRI and PstI
- 3: pSBIC3 mRPS12 TU miniprep 1 cut with PstI
- 4: pSBIC3 mRPS12 TU miniprep 2 cut with PstI
- Gel Order: L, 1, 2, 3, 4
- Gel Results:
- For #2, fragment should be around 500bp
- Gel results indicate that we should redo minipreps or bad enzymes
6/23/16
- (XX) Goal:
- Test if the enzymes are still okay to work with
- Redo miniprep at the same time
- Major steps:
- Enzyme test:
- Tube 1: EcoRI-HF
- Tube 2: PstI-HF
- Tube 3: SpeI-HF
- Tube 4: EarI
- Gel Order: 1, 2, 3, 4, L
- Gel Results: enzymes are working properly
6/24/16
- (HC) After extracting P413 GPD fragment from the gel, I precipitated the DNA
- Precipitating DNA:
- 1.)Add 1-2 μL of 5M NaCl to 30 μL of DNA
- 2.)Add 62 μL of ice cold EtOH
- 3.)Let sit on ice for 30 minutes
- 4.)Centrifuge @ 0℃ for 10 minutes at 13 krpm
- 5.)Remove supernatent
- 6.)Fill tube to halfway point with 70% EtOH
- 7.)Centrifuge @ 4℃ for 2 minutes
- 8.)Remove supernant
- 9.)Let dry
- 10.)Add 6μL of Sterile Water.
- Obtained concentration of DNA with NanoDropper
| Nucleic Acid Concentratione | A @ 260 nm | A @ 280 nm | 260/280 | 260/230 |
|---|---|---|---|---|
| 75.7 ng/μL | 1.514 | 0.922 | 1.64 | .033 |
- (JM/CH)
-
Today we are starting the day off by running a gel.
- We just called iGEM to confirm that we can indeed submit vectors as parts
- The gel we are running today is to confirm that we only have 1 PstI site in all of our plasmids
- The hope: to see one solid band from all of our plasmids except for the controls, the controls should be two bands because they still have two PstI sites
- Another thing we are doing today is we are making media LB Chloro.
- Our Gel:
| 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| 416 control | 426 control | 416 CYC1 | 416 CYC1 | 426 GPD | 426 GPD |
- (XX) Talked about light switch design
- LexA promoter instread of Gal promoter
- PhyB/PIF3 red light system
6/25/16
- (XX) Miniprep mRPS12 TU and mRSP12 mls(total of 8 minipreps)
6/27/16
- (XX) Goal: Redo digest and harvest TU and mls
- Major steps:
- Cut mRPS12 TU with EcoRI/PstI
- Cut Mrps12 mls with EarI/SpeI
- Gel order: L,TU1a, TU1b, TU2a, TU2b, mls1a, mls1b, mls2a, mls2b
- Gel results:
- harvested TU bands from TU2a and TU2b (2 bands per tube)
- TU gel extract = 0.363 g
- harvested four mls bands (2 bands per tube)
- mls gel extract A = 0.342 g
- mls gel extract B = 0.255 g
- Other activities:
- QIA gel extraction kit
- Used nanodrop to test DNA amount:
- TU = 3.1 ng/
- mls A = 41.2 ng/
- mls B = 27.7 ng/
- *according to nanodrop readings, TU needs to be precipitate while we run NEBuilder on mls A and mls B

