| Line 238: | Line 238: | ||
<p style = "font-size:150%; padding:5px 150px 0px 150px; color:#0071A7;">We collaborated with Worcester Polytechnic Institute’s iGEM team to validate the results of our circuit using fluorescent microscopy. The microscope images can be seen below.</p> | <p style = "font-size:150%; padding:5px 150px 0px 150px; color:#0071A7;">We collaborated with Worcester Polytechnic Institute’s iGEM team to validate the results of our circuit using fluorescent microscopy. The microscope images can be seen below.</p> | ||
| − | <center><img src = "https://static.igem.org/mediawiki/2016/thumb/0/08/T--BostonU--WPIMicroscope.png/800px-T--BostonU--WPIMicroscope.png" style = "padding:0px 0px 0px 0px; width: | + | <center><img src = "https://static.igem.org/mediawiki/2016/thumb/0/08/T--BostonU--WPIMicroscope.png/800px-T--BostonU--WPIMicroscope.png" style = "padding:0px 0px 0px 0px; width:80%;"></center> |
<p style = "font-size:150%; padding:0px 150px 15px 150px; color:#0071A7;">The fluorescent microscopy images validated our flow cytometry analysis by showing significant GFP expression in the first state and no significant GFP expression in the other three states.</p> | <p style = "font-size:150%; padding:0px 150px 15px 150px; color:#0071A7;">The fluorescent microscopy images validated our flow cytometry analysis by showing significant GFP expression in the first state and no significant GFP expression in the other three states.</p> | ||
Revision as of 18:13, 16 October 2016
Developing a Digital Parts Library
The first phase of our project was to develop a library of "digital" parts. Our journey began with finding the guide RNA (gRNA) sequences we would use in our work. We generated over one thousand 20 base gRNA sequences in silico using a random sequence generator.
It was critical these gRNA sequences be orthogonal to the human genome because we wanted to test our system in HEK293FT cells and prevent off target activation in the host genome. To test for orthogonality, we entered the sequences into the CRISPR Optimized Design tooldeveloped by the Feng Zhang lab. We selected the top 18 sequences, which had an orthogonality score of 98% or higher, to synthesize and use in our research.

We synthesized these gRNA sequences through IDT and cloned each one into gRNA expressing vectors and gRNA operator pairs. These 18 initial operators expressed an iRFP gene.

We transiently transfected and ran through a flow cytometer our paired gRNA expression vectors and gRNA operator reporters and dCas9-VPR to test their expression behaviors. We wanted to see high activated states coupled with low basal expression, a true "digital" system. The screen of this can be seen below.

Based on this screen, we decided to proceed with gRNA 1 (g1), gRNA 3 (g3), gRNA 8 (g8), and gRNA 13 (g13) for all future experiments. For proper regulation they are paired with the corresponding target sequences g1-Op, g3-Op, g8-Op, and g13-Op.

Next, we needed to prove that our parts could drive a diverse library of genes of interest. To complete this, we replaced the iRFP in our gRNA operator reporters with a GFP, a BFP, and an mRuby protein. The results from these test can be seen in the matrix below.

In all cases, there was low basal expression and strong activation, proving that our system not only behaves "digitally”, but that it could also do so over multiple genes.
Finally, our system needed to be mutually orthogonal to prevent significant undesired crosstalk. If there was off target gene activation between operators, then we could not transfect multiple operator-expression pairs and obtain predictable results. We performed an orthogonality transfection with the GFP driven operators from our library. The experiment was designed such that every gRNA expressing vector would be paired with each gRNA operator reporter vector. We predicted significant GFP fluorescence when the gRNA expressing vector has the same gRNA sequence as the gRNA operator reporter vector, and no significant expression elsewhere. The results of the experiment can be seen in the matrix below.

The diagonal down the matrix demonstrates that there is mutual orthogonality between the gRNAs in our system. With this matrix, we proved that we developed a "digital" parts library.
Our final test was to compare the relative strength of our parts to a CMV (Part:BBa_I712004), a strong mammalian promoter, to determine the relative strength of our system. The graph below shows the results from this experiment.

While our parts expressed well, they did not have a higher expression than the CMV.. But it did raise the question, would we ever be able to show higher activity with a minimal CMV in our system than with a full CMV?
Our next step was to expand to a functioning analog library...
Expanding to an Analog Parts Library
The next step in developing our library was to expand into “analog” expression. We wanted to achieve graded levels of expression.

To create this “digitized-analog” pattern, we had two driving hypotheses: multimerizing the gRNA target sequences would increase expression and mutating the gRNA target sequences would decrease expression.

The concept behind multimerizing, or adding multiple binding sites for the dCas9, harkens to previous work done with various DNA binding domains including ZF (Isalan, Choo, & Klug, 1997), TALE (Bogdanove, 2014), and dCas9 itself (Zhang, et al., 2015). By adding additional binding sites, we expect to recruit more dCas9-VPR transactivators to the gRNA operator reporter vector, increasing expression and producing a synergistically. Additionally, we varied the space between binding sites. We wanted to test if dCas9-VPR complexes bound more effectively and without possible steric hinderance if given more space. The results for experiments for each of the four gRNAs in our library can be seen in the graphs below.

In general, increasing the number of binding sites also increased the expression of the operator reporter. Additionally, with only minor variations from this trend, increasing the space between binding sites also increased expression. This set of experiments proves that multimerization is an effective technique to increase gene activity.
We also compared the relative strength of a CMV (Part:BBa_I712004) from the Registry to all four of our best triple multimerized operator reporters. The data from this comparison can be seen below.
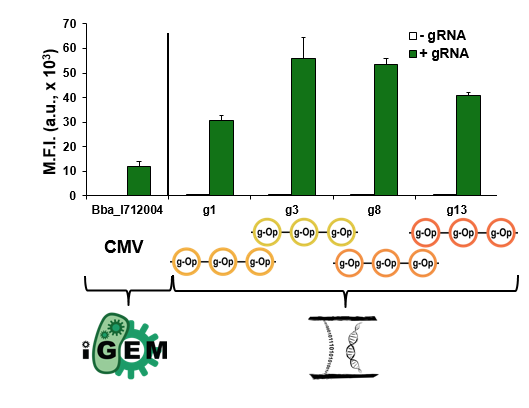
As the graph demonstrates, our triple multimerized operator reporters containing minimal CMVs had greater expression than the full CMV. These parts are broadly useful in other contexts to achieve strong expression in mammalian cells.
After we completed cloning and assaying our multimerized operator reporters, we wanted to develop mutated operator reporters to achieve lower expression levels. We sequentially mutated every nucleotide in our 18 20-nt gRNA target sequences, beginning at the 5’ end. Adenines were mutated to cytosines, and vice versa, and guanines were mutated to thymines, and vice versa. We did this to completely interchange purines and pyrimidines as well as opposite DNA pairs.

Our results showed that we could achieve lower expression levels using mutated operators sites. In particular, mutations at nucleotide 10 and nucleotide 11 produced nearly a fivefold decrease in expression when compared to the non-mutated operator.
In addition to these single nucleotide mutations, we also clustered double and triple point mutations around nucleotide 1 or nucleotide 11 in the gRNA target sequence. The results of this screen can be seen below.

From our multi point mutation experiment, we did also achieve significantly lower expression using the mutations at nucleotide 11.
With these experiment we proved that we developed an "analog" parts library. We have a system than can range from five times below baseline single gRNA target expression to five times above gRNA target expression. Now, what can we accomplish with our library of both digital and "analog" parts?
An answer lies in Genetic Logic Circuits...
Bogdanove, A. J. (2014). Principles and applications of TAL effectors for plant physiology and metabolism. Current Opinion in Plant Biology, 19, 99-104. doi:10.1016/j.pbi.2014.05.007
Isalan, M., Choo, Y., & Klug, A. (1997). Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proceedings of the National Academy of Sciences, 94(11), 5617-5621. doi:10.1073/pnas.94.11.5617
Zhang, Y., Yin, C., Zhang, T., Li, F., Yang, W., Kaminski, R., . . . Hu, W. (2015). CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. Scientific Reports, 5, 16277. doi:10.1038/srep16277
Integration into Genetic Logic Circuits
As we move to combat increasingly complicated problems in synthetic biology, we must create increasingly advanced tools. As synthetic biologists, one route we can take is to develop genetic logic circuits to produce the computational tools needed to complete our research.
In combinational logic design, introducing different combinations of inputs yields different states of output gene expression. Our gene circuits utilize site-specific DNA recombinases to encode logic into cells through DNA rearrangement between specific DNA sequences called recombination sites. Recombinases are powerful because they yield highly digital responses and are efficient in virtually all organisms they have been tested in. Moreover, sophisticated logic functions can be encoded by utilizing heterospecific recombination sites which are mutated sites that efficiently respond to the recombinase but are mutually orthogonal to each other.
We generated an excision-based recombinase reporter design utilized in our lab that expresses a specific gRNA per each combination of two input recombinases. This reporter utilizes two popular tyrosine recombinase widely used in animal genetics. The first, Cre recombinase, excises DNA between two loxP (represented by the pink rectangle like shape in the image below).recombination sites when pointing in the same direction. We used an additional set of sites, called lox2272 (represented by the pink triangle in the image below), which are orthogonal to loxP. The second, Flp recombinase, recognizes sites called FRT (represented by the purple triangles in the image below). Without the presence of Cre or Flp, gRNA 3 is expressed through the constitutive human RNA polymerase III promoter hU6. When Cre is present, the DNA sequences between the lox2272 and loxP sites are deleted and only gRNA 1 is expressed. When Flp is present, the DNA sequence between the FRT sites is removed and only gRNA 8 is expressed. Finally, when Cre and Flp are both present, DNA recombination yields expression of only gRNA 13.

For final production of the desired genes of interest, the gRNAs expressed by the recombinase reporter can activate the synthetic promoters in the Gemini parts library to achieve both digital-to-digital and digital-to-analog responses of output genes. This idea is powerful because the final output gene expression is decoupled from the encoding of the logic on the recombinase reporter. Therefore, a host of different functions can be encoded without having to build a new recombinase reporter vector once a library of synthetic promoters and output genes is established.
Experiment 1: Single “digital” output functions
We sought to first achieve a simple logic function, called a 2-input AND gate. This function yields an output when only both inputs are expressed. We transfected our logic circuit and a guide RNA operator containing a g13 target site and a BFP reporter into HEK293FT cells. With this design, we expected that there would be strong BFP expression in state 4 (+Cre, +Flp) but no significant fluorescence in states 1, 2, or 3. As the graph below demonstrates, we succeeded in proving our system could control gene activation via an AND gate.

A NOR gate will only express a gene in the absence of signals. We developed a NOR gate circuit and transfected it into our HEK cells along with a guide RNA operator containing a g3 target site and a GFP reporter. We expected to see significant GFP expression only in state 1 (-Cre, -Flp) and no significant expression in the other three states. As the graph below demonstrates, we succeeded in proving our system could work effectively in a NOR gate to repress a desired gene.

We collaborated with Worcester Polytechnic Institute’s iGEM team to validate the results of our circuit using fluorescent microscopy. The microscope images can be seen below.

The fluorescent microscopy images validated our flow cytometry analysis by showing significant GFP expression in the first state and no significant GFP expression in the other three states.
Experiment 2: Multiple “digital” output functions.
After proving our library’s ability to perform fundamental circuits behaviors, we developed more complex behaviors. We combined our AND and NOR gates to develop a multi-color XNOR functionality. We expected to see GFP expression in the absence of signals in our first state, no significant fluorescence in our second and third states, and BFP expression in the presence of both signals in our fourth state. To perform this experiment, we transfected our circuit and the operators from our previous AND and NOR gate experiments into HEK cells and assayed fluorescence using our flow cytometers. As the results below show, our circuit functions as expected, proving our circuits capacity to perform more complex functions than fundamental single-output.

For our final "digital" output circuit, we developed line decoders. A line decoder is when each logical state has its own output. We created four distinct line decoders with each state producing a different fluorescent protein (BFP, GFP, iRFP, and mRuby) in each decoder. The results of these four trials can be seen below with the operators used in the experiment stated:
g3- BFP g1- GFP g8-iRFP g13- mRuby

g3- iRFP g1- mRuby g8- BFP g13- GFP

g3- mRuby g1- BFP g8- GFP g13-iRFP

g3- GFP g1- iRFP g8- mRuby g13- BFP

In each line decoder, there is only significant protein fluorescence in its predicted state, proving that our library is fully capable of performing in complex systems composed of two inputs and four outputs.
Experiment 3: Graded “analog” output responses.
Once we were able to prove our system could produce "digital" output, we tested to see if we could also produce a circuit that could provide "analog" outputs. An "analog" output circuit has the same gene of interest expressed at different levels for a given logical state. In our circuit, state 1 paired with a triple multimerized operator reporter with a g3 target site. State 2 paired with a single binding site operator with a g1 target site. State 3 paired with the double multimerized operator reporter with a g8 target site. Finally, state 4 paired with a mutant operator with a g13 target site. All of these operators contained a GFP reporter and were transfected with the genetic logic circuit and their accompanied expression vectors and dCas9-VPR. We predicted that the order of expression, from highest to lowest, would be state 1, state 3, state 2, and finally state 4. The results of the experiment can be seen in the graphic below.

While there are some discrepancies between the expected results and the actual results, (specifically the expression level of state 3), we can say that this experiment effectively proves that the Gemini library of parts can be used to produce an "analog" response from a combinatorial circuit.


