Introduction

Higher order self-assembling protein assemblies are commonplace in nature, such as ferritin, which carries iron in many single-celled organisms. Due to their encapsulating function and box-like structure, such assemblies are often called protein “cages.” Inspired by these natural sources, an abundance of research has been done into creating synthetic cages with new customized properties such as stability, size, and subunit types. Furthermore, while applications for cages have been suggested, many have not been tested experimentally.
An application that we have centered our project around is targeted drug delivery. By specifying the site of delivery, the effects of the drug can be limited to desired areas with maximized efficiency and we can avoid damaging undesired targets in the body.
Thrombosis, the process by which a clot forms in the blood stream, is associated with widespread diseases including stroke and many heart conditions. Anticoagulants prevent clot formation by interfering with clotting factors and allowing for smoother blood flow and restoring normal circulation. However, these drugs can have detrimental side effects, such as an inability to inhibit fatal blood loss through cuts or other wounds. By localizing the site of delivery and reducing the necessary drug dosage, we can mitigate these kinds of problems. A protein cage encapsulating an anticoagulant drug that would selectively release its payload at the site of a clot would fulfill this purpose. To specify the target, we should select an associated enzyme or byproduct of the clotting cascade. Thrombin, a protease present at blood clots, cleaves a specific amino acid sequence. This sequence, if properly inserted into a protein cage containing an anticoagulant, would result in disassembly of the cage, release of the drug molecule to the targeted area, and destruction of the clot.
The goal of our project is to modify protein cages that disassemble in the presence of thrombin protease. We are using two previously created synthetic cages, one composed of 12 subunits forming a tetrahedral cage while the other is composed of 24 subunits forming an octahedral cage. For each of these protein cages, we have designed mutants with inserted thrombin cleavage sites with the intention to induce disassembly of the cages by treatment with thrombin protease. Future directions of this project consist of loading anticoagulant drug molecules into our mutant cages and assaying for the release of the drug molecules upon disassembly in the presence of thrombin.
Methodology
The main goal of the project is to develop a mechanism for targeted drug delivery. By incorporating protease cleavage sites into our protein cages, we control disassembly of the cage simply by providing it with a specific protease. The protease we decided to work with is Thrombin, a crucial member in the blood clotting cascade. Its role in Thrombosis makes it a key target in many of our leading causes of death including, heart attacks and strokes. By incorporating Thrombin cleavage sites into our cage and using Thrombin as our targeting molecule, we hope to directly combat heart attacks and strokes using our drug delivery mechanism.
The design of the project involves three distinct steps.
1. The first step revolves around the design of our mutants derived from the wild type cage. Because of the sensitive nature of these protein cages, there were specific criteria that we had set to maximize mutant formation and guarantee disassembly of our cage once it came into contact with the protease
- Design mutations away from any possibly secondary or tertiary structures.-The main reasoning behind this criteria is to avoid disruption of the protein cage. It is difficult to gauge which sequences are crucial in cage formation so by avoiding mutations in secondary or tertiary structures, we hoped to mutate a region that would have a minimal impact in cage formation and thus cage disruption.
- Design mutations on the exterior of the cage.-The main reasoning behind this criteria is to allow for easy access by the Thrombin protease. By placing the mutation on the exterior of the cage, we hoped to reduce steric hindrance to the protease and maximize chances of cleavage.
- Design mutations near the linker region (ONLY FOR PC Quad)-The main reasoning behind this criteria is to help disassemble the cage. In the original design of the wild type PCquad cage, it is understood that the there is a linker region that serves as the backbone of a subunit thus proving crucial to the cage structure. By mutating a region close to the linker, we hoped to maximize chances of cage disassembly.
Using these criteria, we designed 10 mutant cages for PCquad and 3 mutant cages for O3-33.
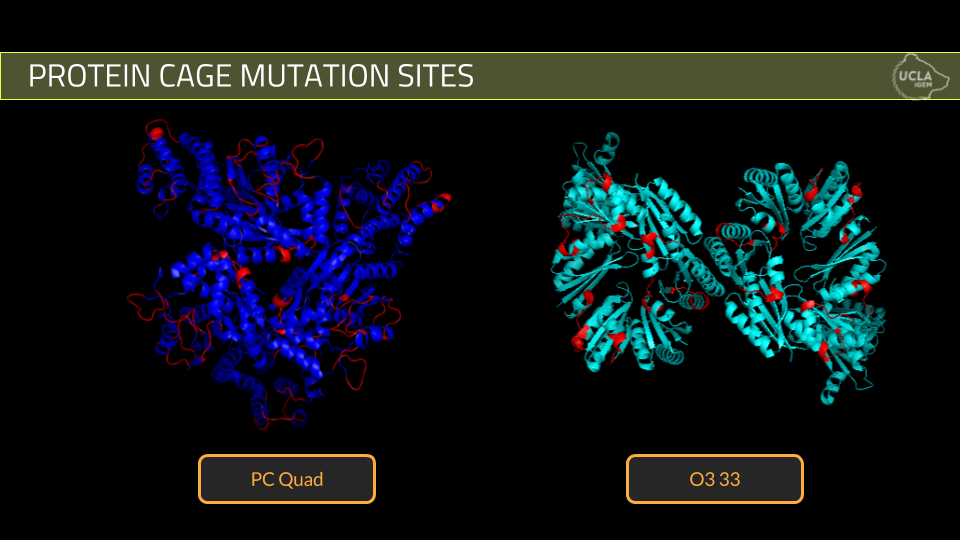
2. The second step of the project is protein expression and verification of cage formation. We attempted expression of five of our best PCquad designs and all three of our O3-33 designs. Expression was done using standard expression protocols and purification was done using his -tag nickel column purification while cage formation was verified using Dynamic Light Scattering.
- Dynamic light scattering is used to determine the approximate size and polydispersity of small particles in solution. A beam of light is shown through the sample and the fluctuations in the light due to the movement of the small particles is measured. DLS is dependent on size to the fourth order so it is very important to filter out any large particles such as dust before conducting the experiment.
- In our experiment, DLS was used to determine whether or not our protein cages formed as well as the purity of our samples. A radius around 8 nm for PCQuad (WT +mutants) and a radius around 6.5 nm for O3-33 (WT + mutants) as well as a polydispersity below 30%, indicated by DLS meant a pure sample.
3. The final step of the project involved testing our successfully formed mutant cages for disassembly in the presence of Thrombin protease. The mutant cages that had successfully formed cages were treated to Thrombin protease at various concentrations and durations. Disassembly of cage was verified using a combination of SDS page to see if the individual subunits cut at the locations of the cleavage site insert and DLS to see if the entire cage structure fell apart.
General Protocol
- 60 uL 0.2 mg/ml protein cage samples used for thrombin assay
- For each protein cage tested, a sample was subject to thrombin while another was not
- Added 1.25 uL of 460 units/ml bovine thrombin stock to each sample (0.69 units) to each sample that was to be subject to thrombin every 4 for hours for 16 hours
- An SDS-PAGE gel was run to assay for cleavage and a DLS test was conducted to assay for protein cage disassembly
For a list and description of all protocols use, visit our Protocols page here.
Results
Overview
We have successfully been able to incorporate a thrombin protease cleavage site onto the exterior of two different protein cage nanostructures and induce disassembly of these cages upon treatment with a thrombin protease. The results of our project demonstrate that mutations can be inserted into protein cages in order to modify them into potential targeted drug delivery vehicles. Future plans include loading these protein cages with various small molecules and assaying for exposure to the external environment upon cleavage and disassembly.
Cloning
All PCQuad and O3-33 wildtype and mutant protein cages were successfully cloned into psb1c3 vector as biobricks, sequence verified and submitted to the parts registry. As shown in Figure 1, the biobricks contain a T7 promoter, Ribosome Binding Site, sequence for an individual subunit of the specific cage, a 6x Histadine tag and a stop codon.

Figure 1. Sequence for PCQuad Mutant M1.
Expression and Purification
PCQuad and O3-33 Wild Type protein cage subunits were successfully expressed and purified; this was confirmed via SDS-PAGE.

Figure 2. SDS-PAGE for expression of PCQuad and O3-33 WildType Protein Cages.The size of a subunit of PCQuad is approximately 50 kDa and the size of a subunit of O3-33 is approximately 15 kDa.
PCQuad Mutant M1 and Mutant M13 subunits were successfully expressed and purified. PCQuad Mutant M2, Mutant M4 and Mutant M10 subunits were not able to be purified.

O3-33 Mutant aa88 subunit was successfully expressed and purified. O3-33 Mutant aa118 and Mutant aa173 subunits were not able to be purified.

Cage Formation
Dynamic light scattering was used to determine whether the expressed subunits self-assembled into the protein cage structure. A radius of around 8 nm is expected for PCQuad WIldType and its mutants and a radius of around 6.5 nm is expected for O3-33 and its mutants. A polydispersity reading below 30% is desired to ensure that our substance is pure and specific.
PCQuad and O3-33 WildType subunits successfully self-assembled into protein cage structures.

DLS Results for PCQuad WildType Cage and O3-33 WildType Cage
PCQuad Mutant M1 and O3-33 Mutant aa88 subunits successfully self-assembled into protein cage structures. PCQuat Mutant M13 subunits did not.

DLS Results for PCQuad Mutant M1 Cage, PCQuad Mutant M13 Cage, and O3-33 Mutant aa88 Cage
Thrombin Assay
Results from the Thrombin Assay successfully demonstrate cleavage of individual subunits and cage disassembly of PCQuad Mutant M1 and O3-33 Mutant aa88. WildType PCQUad and O3-33 protein cages were unaffected in the presence of thrombin.

DLS Results for PCQuad Mutant M1 before addition of thrombin and after addition of thrombin

DLS Results for O3-33 Mutant aa88 before addition of thrombin and after addition of thrombin
We have strong evidence that our project works! To see our Proof of Concept (Gold Medal Criterion), please click here.
Biobricks
- BBa_K2068000
- BBa_K2068001
- BBa_K2068002
- BBa_K2068003
- BBa_K2068004
- BBa_K2068005
- BBa_K2068006
- BBa_K2068007
- BBa_K2068008






