|
|
| Line 91: |
Line 91: |
| | <p><li>Synechococcus sp PCC 7942 had no signals of life.</li><br /> | | <p><li>Synechococcus sp PCC 7942 had no signals of life.</li><br /> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" alt="IMG_2956" width="300px"> | + | <img align="center" src="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" alt="Igem-6803-week2" width="300px"> |
| | </a> | | </a> |
| | | | |
| Line 103: |
Line 103: |
| | <h1 class="entry-title">Week3(8/29/2016-9/4/2016)</h1> | | <h1 class="entry-title">Week3(8/29/2016-9/4/2016)</h1> |
| | <div class="entry-content"> | | <div class="entry-content"> |
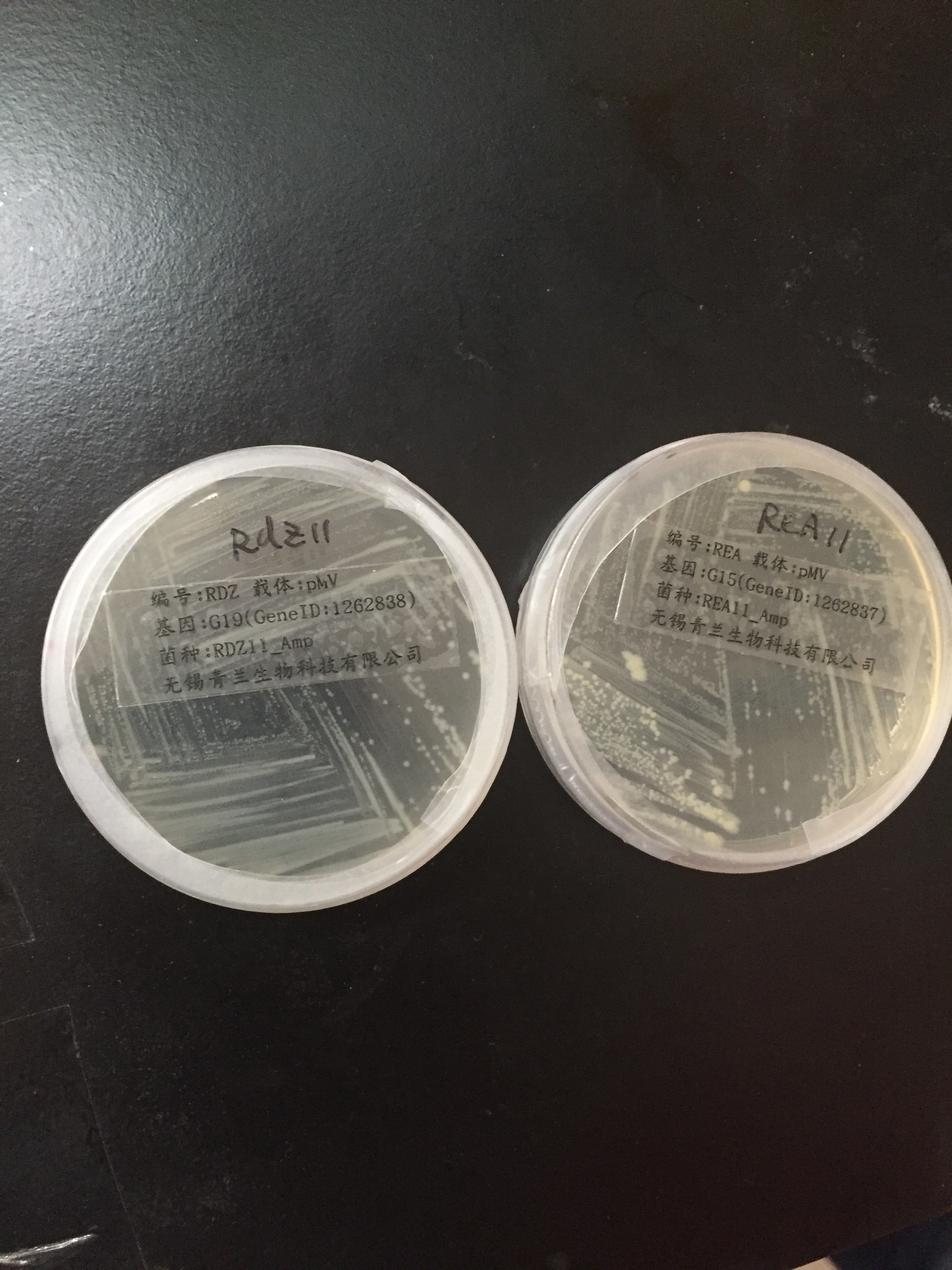
| − | <p><li>pMV-G19 and pMV-G15 containing our target genes were received.</li> | + | <p><li>pMV-G19 and pMV-G15 containing our target genes were received.</li><br/> |
| | | | |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" alt="IMG_2956" width="300px" > | + | <img align="center" src="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" alt="Igem-6803-week3-1" width="300px" > |
| | </a> | | </a> |
| | | | |
| Line 154: |
Line 154: |
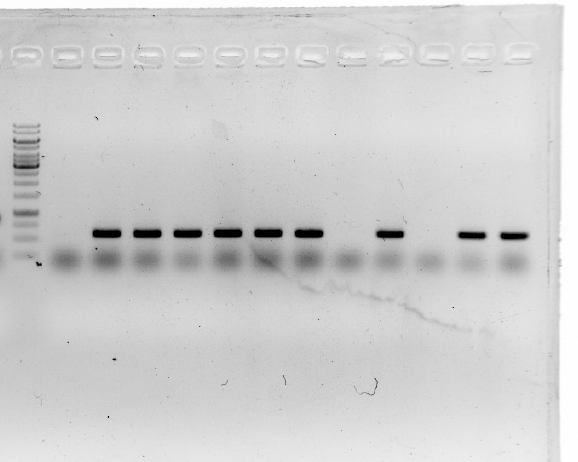
| | <li>Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.</li> | | <li>Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.</li> |
| | <li>Mono-restriction digest of pT-19-15 with Nru I.</li> | | <li>Mono-restriction digest of pT-19-15 with Nru I.</li> |
| − | <li>The enzyme-digested product was dephosphorylation.</li> | + | <li>The enzyme-digested product was dephosphorylation.</li><br/> |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="IMG_2956" width="800" height="533" | + | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="800" height="533" |
| | </a> | | </a> |
| | | | |
| Line 207: |
Line 207: |
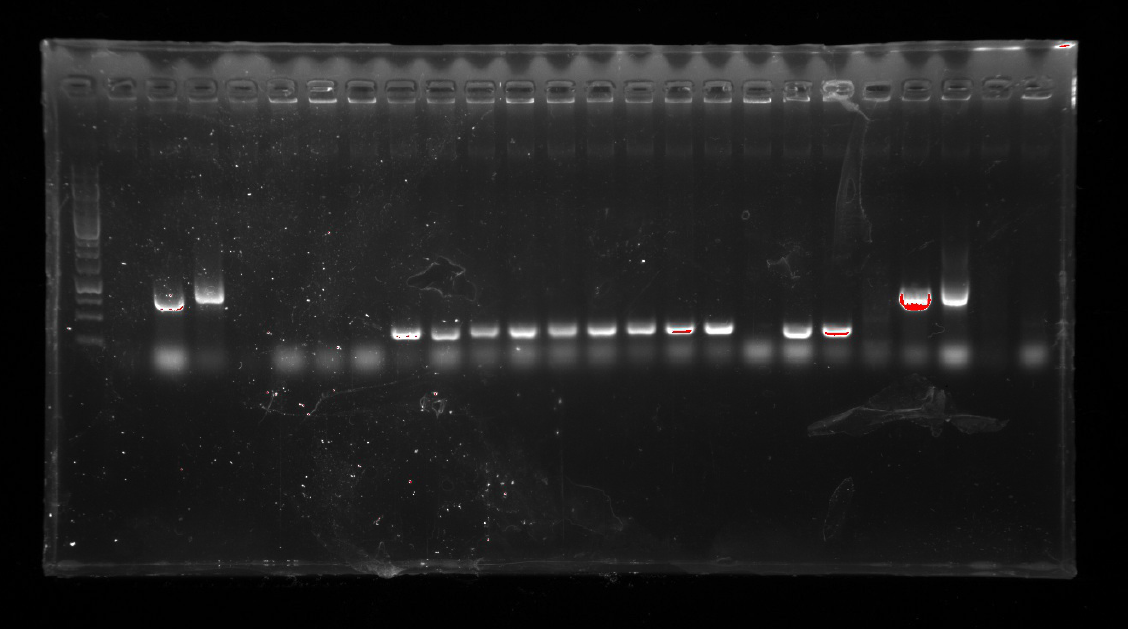
| | <li>A colony PCR of pT-13-19-15 was performed with 7 colonies.</li> | | <li>A colony PCR of pT-13-19-15 was performed with 7 colonies.</li> |
| | Two of the successful ones were used to inoculate overnight cultures.</li> | | Two of the successful ones were used to inoculate overnight cultures.</li> |
| − | <li>Two kinds of plasmids were isolated using a miniprep kit.</li> | + | <li>Two kinds of plasmids were isolated using a miniprep kit.</li><br/> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="IMG_2956" width="800" height="533"> | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="800" height="533"> |
| | </a> | | </a> |
| | | | |
| Line 248: |
Line 248: |
| | 2.13_fwd and 19_rev on pT-13-19-15<br/> | | 2.13_fwd and 19_rev on pT-13-19-15<br/> |
| | <b>The second one was failed.</b><br/> | | <b>The second one was failed.</b><br/> |
| − | <img align="center"src="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" alt="IMG_2956" width="800" height="533" > | + | <img align="center"src="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" alt="Igem-6803-week5" width="800" height="533" > |
| | </a> | | </a> |
| | | | |
| Line 284: |
Line 284: |
| | <li>Ligated <i>13</i> and <i>19-15</i> via overlap PCR.</li> | | <li>Ligated <i>13</i> and <i>19-15</i> via overlap PCR.</li> |
| | This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.</li> | | This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.</li> |
| − | <li>Restriction digest on pCPC-3031-Ni with Sac I.</li> | + | <li>Restriction digest on pCPC-3031-Ni with Sac I.</li><br/> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="IMG_2956" width="300px"> | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"> |
| | | | |
| − | <li>The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.</li> | + | <li>The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.</li><br/> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="IMG_2956" width="300px" > | + | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" > |
| | | | |
| | <li>A colony PCR of pT-13-19-15 was performed with 12 colonies.</li> | | <li>A colony PCR of pT-13-19-15 was performed with 12 colonies.</li> |
| Line 299: |
Line 299: |
| | <li>Plasmids pT-13-19-15 were isolated using a miniprep kit.</li> | | <li>Plasmids pT-13-19-15 were isolated using a miniprep kit.</li> |
| | <li>A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.</li> | | <li>A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.</li> |
| − | Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.</li> | + | Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.</li><br/> |
| | | | |
| | | | |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="IMG_2956" width="800" height="533" > | + | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" > |
| | | | |
| | | | |