Crepuscule (Talk | contribs) (Created page with "{{:Team:Tianjin/Templates/MaterialTheme|}} {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/R-R/style.css}} {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Community/...") |
|||
| (54 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
{{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/R-R/style.css}} | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/R-R/style.css}} | ||
{{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Community/css/bootstrap.css}} | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Community/css/bootstrap.css}} | ||
| − | |||
| − | |||
| − | |||
<html> | <html> | ||
| Line 44: | Line 41: | ||
<!-- <div class="row"> --> | <!-- <div class="row"> --> | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | < | + | <h2 id="about" class="title text-center">Experiment of <span>A Controllable Lipid Producer</span></h2> |
<h2><b>Overview</b></h2> | <h2><b>Overview</b></h2> | ||
| − | <p style="font-size:18px"> | + | <p style="font-size:18px">Cyanobacteria are excellent organisms for biofuel production. We thus have selected Cyanobacterium <i>Synechocystis sp. PCC 6803</i> as the source of carbon in our mixed bacteria system. Our target is simply to make the cyanobacteria lyse at the appropriate time by transforming a plasmid contained three bacteriophage-derived lysis genes which were placed downstream of a nickel-inducible signal transduction system into the <i>Synechocystis 6803</i>.</p> |
| Line 52: | Line 49: | ||
<div class="col-md-3"> | <div class="col-md-3"> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/9/9f/Igem-6803-e1.png" alt="desktop"> |
| − | + | <p style="font-size:15px"> Fig.1. Lipid contents</p> | |
| − | + | ||
</div> | </div> | ||
<div class="col-md-9"> | <div class="col-md-9"> | ||
| − | <h3><b>1. | + | <h3><b id="LipidProducer">1. Lipid Producer</b></h3> |
| − | <p style="font-size:18px"> | + | <p style="font-size:18px">Photosynthetic microorganisms, including eukaryotic algae and cyanobacteria, are being optimized to overproduce numerous biofuel. According to previous data, algae accumulate large quantities of lipid as storage materials, but they do this when under stress and growing slowly. By contrast, cyanobacteria accumulate lipids in thylakoid membranes, which are associated with high levels of photosynthesis and a rapid growth rate. Thus, photo-synthetic bacteria have a natural advantage for producing lipids at a high rate. Furthermore, being prokaryotes can be improved by genetic manipulations much more readily than can eukaryotic algae. (Espaux L et al. 2015) Therefore, we decided to do something to make cyanobacteria ,the lipid producer, more appropriate for our project.</p> |
</div> | </div> | ||
| Line 67: | Line 64: | ||
| − | <div class="col-md- | + | <div class="col-md-12"> |
| + | |||
| + | <p style="font-size:18px"><i>Synechococcus elongatus PCC7942</i> has larger capacity of lipid production than <i>Synechocystis sp. PCC6803</i> but accumulates most of the product in the cell because of the imbalance of the rates of lipid production and secretion. Initially, we intended to do something to increase lipid secretion by knocking the <i>wzt</i> gene(Akihiro Kato et al. 2016), however, <i>Synechococcus elongatus PCC7942</i> wasn’t able to revive in two-week shaking cultivation. So we turned into <i>Synechocystis sp.PCC 6803</i>.</p> | ||
| + | |||
| + | |||
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="col-md-2"></div> | <div class="col-md-2"></div> | ||
| Line 85: | Line 82: | ||
<div class="row"> | <div class="row"> | ||
<div class="col-md-5"> | <div class="col-md-5"> | ||
| − | <h3><b>2. | + | <h3><b class="LipidRecoveryFromBiomass">2.Lipid Recovery From Biomass</b></h3> |
| − | <br/> <p style="font-size:18px">The | + | <br/> <p style="font-size:18px">The first goal of our research was to facilitate lipid recovery from biomass. The scientific community widespread disrupts the cyanobacterial cell envelope to achieve the goal. (Seog JL et al. 1998)However, all these methods are not economical for large amounts of biomass or add additional cost and reduce the overall utility of the process. Our target is simply to make the cyanobacteria lyse at the appropriate time.</br> |
| + | We found that the cyanobacterial cell envelope is composed of 4 layers: the external surface layers ;the outer membrane; the polypeptidoglycan which is considerably thick, and the cytoplasmic membrane.( Hoiczyk E et al. 2000)To break up the peptidoglycan layer, we applied the holin-endolysin lysis strategy used by bacteriophages to exit bacterial cells(Wang IN et al. 2000). Endolysins are peptidoglycan-degrading enzymes that attack the covalent linkages of the peptidoglycans that maintain the integrity of the cell wall. In addition to endolysins, some auxiliary lysis factors are involved in cleaving the oligopeptide linkages between the peptidoglycan and the outer membrane lipoprotein. Holins are small membrane proteins that produce nonspecific lesions (holes) in the</p> | ||
</div> | </div> | ||
<div class="col-md-7"> | <div class="col-md-7"> | ||
<br/><br/><br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/><br/><br/> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/d/d5/Igem-6803-e2.jpg" alt="desktop"> |
| − | <p style="font-size:15px"> Fig. | + | <p style="font-size:15px"> Fig.2. The functions of holin and endolysins in degrading the cell wall</p> |
| + | <br/><br/><br/> | ||
| + | <p style="font-size:18px"> cytoplasmic membrane from within, allow the endolysins and auxiliary lysis factors to gain access to the polypeptidoglycan layers, and trigger the lysis process. In this way, the cell wall is easy to break up.</p> | ||
| + | |||
| Line 99: | Line 100: | ||
<div class="row"> | <div class="row"> | ||
| − | <div class="col-md- | + | <div class="col-md-12"> |
| − | + | <h3><b id="ControlTheLysisSystem">3. Control The Lysis System</b></h3> | |
| − | + | <p style="font-size:18px"><br/>To control the appropriate time, a nickel sensing/responding signal system(Garcia-Dominguez M et al. 2000) was used to control the timing of the expression of phage lysis genes in Synechocystis 6803.</BR> | |
| − | + | Our strategy for achieving our target is to construct a expression vector pCPC3031-Ni-13-19-15 introduced the Salmonella phage P22 lysis cassette (<I>13-19-15</I>) with a Ampicillin selection marker downstream of the promoter Pni, a nickel responding signal operon. Synechocystis 6803 with the pCPC3031-Ni-13-19-15 will lyse after Ni2+ addition. | |
| − | < | + | |
| − | + | ||
</p> | </p> | ||
| − | |||
| − | |||
| − | |||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
| − | <h2><b> | + | <h2><b>Aim</b></h2> |
<div class="row"> | <div class="row"> | ||
| − | <div class="col-md- | + | <div class="col-md-12"> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | <br/> <p style="font-size:18px">In this part of our project, Cyanobacterium Synechocystis sp. PCC 6803 was selected as a model organism as the source of carbon in our mixed bacteria system. We simply to establish a cell wall disruption process which could make the cyanobacteria lyse at the appropriate time. </p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | + | <div class="col-md-12"> | |
| − | + | ||
| + | <h2><b>Strategies</b></h2> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="row"> | <div class="row"> | ||
| − | <div class="col-md- | + | <div class="col-md-12"> |
| − | <h3><b> | + | <h3><b>1. Lysis genes(<i>13-19-15</i>)</b></h3> |
| − | <br/> <p style="font-size:18px"> | + | <br/> <p style="font-size:18px">P22 gp13,P22 gp19 and P22 gp 15 are holins, endolysins and auxiliary lysis factors respectively. To construct the holin-endolysin lysis system, they should be connected together with defined sequence. First of all, we obtained the three lysis genes which were synthesized by GENEWIZ separately. Then we use PCR to amplify this part. TA cloning and ligation of Blunt-ended DNA on the T vector were our original idea.However, <i>19</i> and <i>15</i> were spliced via TA cloning according to our presumption. <i>13</i> and <i>19-15</i> were ligated PCR overlap extension method of Warrens et al.( Warrens AN et al. 1997)</p> </br></br></br> |
| − | + | ||
| − | + | ||
| − | + | ||
| + | <img src="https://static.igem.org/mediawiki/2016/8/8e/Igem-6803-e3.png" alt="desktop"> | ||
| + | <p style="font-size:15px"> Fig.3. TA cloning and blunt end ligation </p></br></br> | ||
</div> | </div> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<div class="col-md-5"> | <div class="col-md-5"> | ||
| − | |||
| − | |||
| + | <h3><b>2.A Nickel Sensing/Responding Signal System(pCPC3031-Ni)</b></h3><br/> | ||
| + | <p style="font-size:18px">Ni activates the transcription of downstream genes of Pni and positively autoregulates its own synthesis. The amount of mRNA increased about 20-fold within 4 h after Ni addition. First, Pni was cloned into pCPC3031 and was amplified by using PCR. Then we cut the plasmid with Nru I, after that the lysis genes, <I>13-19-15</I>, were placed downstream of Pni. What’ more, we handed in the plasimids for sequencing, which confirmed its correctness.</p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="col-md-7"> | <div class="col-md-7"> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/c/c4/IGEM-6803-E4.png" alt="desktop"> |
| − | <p style="font-size:15px"> Fig. | + | <p style="font-size:15px"> Fig.4. Overlap PCR</p> |
</div> | </div> | ||
| − | |||
| − | <div | + | </div |
| − | + | ||
| − | + | ||
| − | |||
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | <h2><b> | + | <h2><b>Summary</b></h2> |
| − | <p style="font-size:18px"> | + | <p style="font-size:18px">After analyzing and comparing Photosynthetic microorganisms from one to another indetail in two aspects of maneuverability and lipid production, we first chose <i>Synechocystis 6803</i> as the lipid producer in mixed cultivation. To release more production, three genes encoding proteins which could make the cells split were inserted into a plasmid .The next step was to lyse the bacteria at specific time, therefore we put Pni, a promoter, to the upstream of lysis genes. To sum up, we constructed Nickel-inducible lysis system in <i>Synechocystis sp.PCC 6803</i> .</p> |
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<h2><b>References</b></h2> | <h2><b>References</b></h2> | ||
| − | <p style="font-size:16px"><i>[1] | + | <p style="font-size:16px"><i>[1]Espaux L, Mendez-Perez D, Li R, Keasling JD (2015) Synthetic biology for microbial production of lipid-based biofuels. <i>Curr Opin Chem Biol</i>. 29:58-65</br></br> |
| − | [2] | + | [2]Seog JL, Byung-Dae Y, O. H-M (1998) Rapid method for the determination of lipid fromthe green alga Botryococcus braunii.<i> Biotechnol Tech </i>12:553–556.</br></br> |
| − | [3] | + | [3]Hoiczyk E, HanselA(2000) Cyanobacterial cell walls: News from an unusual prokaryotic envelope. <i>J Bacteriol </i>182:1191–1199.</br></br> |
| − | + | [4]Wang IN, Smith DL, Young R (2000) Holins: The protein clocks of bacteriophage infections.<i> Annu Rev Microbiol</i> 54:799–825.</br></br> | |
| − | [ | + | [5]Garcia-Dominguez M, Lopez-Maury L, Florencio FJ, Reyes JC (2000) A gene clusterinvolved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. <i>J Bacteriol</i> 182:1507–1514.</br></br> |
| + | [6]Warrens AN, Jones MD, Lechler RI (1997) Splicing by overlap extension by PCR using asymmetric amplification: An improved technique for the generation of hybrid proteins of immunological interest.<i> Gene</i> 186:29–35.</br></br> | ||
| + | [7]Akihiro Kato, Kazuhide Use, Nobuyuki Takatani, Kazutaka Ikeda, Miyuki Matsuura, Kouji Kojima (2016) Modulation of the balance of fatty acid production and secretion is crucial for enhancement of growth and productivity of the engineered mutant of the cyanobacterium Synechococcus elongates. <i>Biotechnol Biofuels</i>9:91-101.</i></br></br> | ||
| + | </p><br/><br/></br> | ||
</div></div> | </div></div> | ||
| Line 306: | Line 178: | ||
| − | + | </body> | |
</html> | </html> | ||
{{:Team:Tianjin/Templates/Sponsor|}} | {{:Team:Tianjin/Templates/Sponsor|}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 16:11, 15 October 2016
Experiment of A Controllable Lipid Producer
Overview
Cyanobacteria are excellent organisms for biofuel production. We thus have selected Cyanobacterium Synechocystis sp. PCC 6803 as the source of carbon in our mixed bacteria system. Our target is simply to make the cyanobacteria lyse at the appropriate time by transforming a plasmid contained three bacteriophage-derived lysis genes which were placed downstream of a nickel-inducible signal transduction system into the Synechocystis 6803.

Fig.1. Lipid contents
1. Lipid Producer
Photosynthetic microorganisms, including eukaryotic algae and cyanobacteria, are being optimized to overproduce numerous biofuel. According to previous data, algae accumulate large quantities of lipid as storage materials, but they do this when under stress and growing slowly. By contrast, cyanobacteria accumulate lipids in thylakoid membranes, which are associated with high levels of photosynthesis and a rapid growth rate. Thus, photo-synthetic bacteria have a natural advantage for producing lipids at a high rate. Furthermore, being prokaryotes can be improved by genetic manipulations much more readily than can eukaryotic algae. (Espaux L et al. 2015) Therefore, we decided to do something to make cyanobacteria ,the lipid producer, more appropriate for our project.
Synechococcus elongatus PCC7942 has larger capacity of lipid production than Synechocystis sp. PCC6803 but accumulates most of the product in the cell because of the imbalance of the rates of lipid production and secretion. Initially, we intended to do something to increase lipid secretion by knocking the wzt gene(Akihiro Kato et al. 2016), however, Synechococcus elongatus PCC7942 wasn’t able to revive in two-week shaking cultivation. So we turned into Synechocystis sp.PCC 6803.
2.Lipid Recovery From Biomass
The first goal of our research was to facilitate lipid recovery from biomass. The scientific community widespread disrupts the cyanobacterial cell envelope to achieve the goal. (Seog JL et al. 1998)However, all these methods are not economical for large amounts of biomass or add additional cost and reduce the overall utility of the process. Our target is simply to make the cyanobacteria lyse at the appropriate time. We found that the cyanobacterial cell envelope is composed of 4 layers: the external surface layers ;the outer membrane; the polypeptidoglycan which is considerably thick, and the cytoplasmic membrane.( Hoiczyk E et al. 2000)To break up the peptidoglycan layer, we applied the holin-endolysin lysis strategy used by bacteriophages to exit bacterial cells(Wang IN et al. 2000). Endolysins are peptidoglycan-degrading enzymes that attack the covalent linkages of the peptidoglycans that maintain the integrity of the cell wall. In addition to endolysins, some auxiliary lysis factors are involved in cleaving the oligopeptide linkages between the peptidoglycan and the outer membrane lipoprotein. Holins are small membrane proteins that produce nonspecific lesions (holes) in the

Fig.2. The functions of holin and endolysins in degrading the cell wall
cytoplasmic membrane from within, allow the endolysins and auxiliary lysis factors to gain access to the polypeptidoglycan layers, and trigger the lysis process. In this way, the cell wall is easy to break up.
3. Control The Lysis System
To control the appropriate time, a nickel sensing/responding signal system(Garcia-Dominguez M et al. 2000) was used to control the timing of the expression of phage lysis genes in Synechocystis 6803.
Our strategy for achieving our target is to construct a expression vector pCPC3031-Ni-13-19-15 introduced the Salmonella phage P22 lysis cassette (13-19-15) with a Ampicillin selection marker downstream of the promoter Pni, a nickel responding signal operon. Synechocystis 6803 with the pCPC3031-Ni-13-19-15 will lyse after Ni2+ addition.
Aim
In this part of our project, Cyanobacterium Synechocystis sp. PCC 6803 was selected as a model organism as the source of carbon in our mixed bacteria system. We simply to establish a cell wall disruption process which could make the cyanobacteria lyse at the appropriate time.
Strategies
1. Lysis genes(13-19-15)
P22 gp13,P22 gp19 and P22 gp 15 are holins, endolysins and auxiliary lysis factors respectively. To construct the holin-endolysin lysis system, they should be connected together with defined sequence. First of all, we obtained the three lysis genes which were synthesized by GENEWIZ separately. Then we use PCR to amplify this part. TA cloning and ligation of Blunt-ended DNA on the T vector were our original idea.However, 19 and 15 were spliced via TA cloning according to our presumption. 13 and 19-15 were ligated PCR overlap extension method of Warrens et al.( Warrens AN et al. 1997)

Fig.3. TA cloning and blunt end ligation
2.A Nickel Sensing/Responding Signal System(pCPC3031-Ni)
Ni activates the transcription of downstream genes of Pni and positively autoregulates its own synthesis. The amount of mRNA increased about 20-fold within 4 h after Ni addition. First, Pni was cloned into pCPC3031 and was amplified by using PCR. Then we cut the plasmid with Nru I, after that the lysis genes, 13-19-15, were placed downstream of Pni. What’ more, we handed in the plasimids for sequencing, which confirmed its correctness.
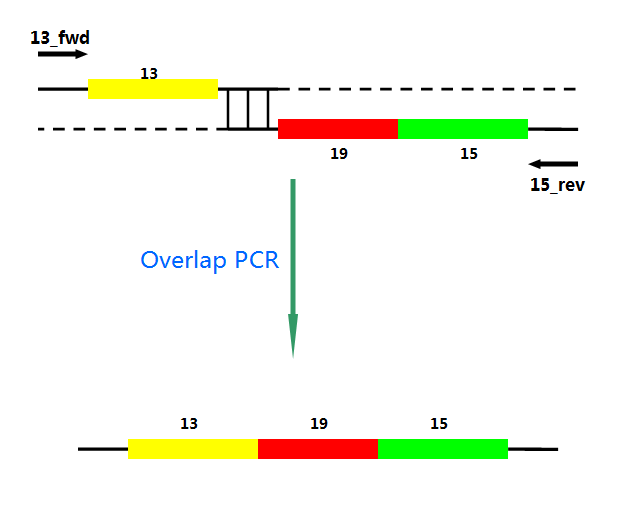
Fig.4. Overlap PCR
Summary
After analyzing and comparing Photosynthetic microorganisms from one to another indetail in two aspects of maneuverability and lipid production, we first chose Synechocystis 6803 as the lipid producer in mixed cultivation. To release more production, three genes encoding proteins which could make the cells split were inserted into a plasmid .The next step was to lyse the bacteria at specific time, therefore we put Pni, a promoter, to the upstream of lysis genes. To sum up, we constructed Nickel-inducible lysis system in Synechocystis sp.PCC 6803 .
References
[1]Espaux L, Mendez-Perez D, Li R, Keasling JD (2015) Synthetic biology for microbial production of lipid-based biofuels. Curr Opin Chem Biol. 29:58-65 [2]Seog JL, Byung-Dae Y, O. H-M (1998) Rapid method for the determination of lipid fromthe green alga Botryococcus braunii. Biotechnol Tech 12:553–556. [3]Hoiczyk E, HanselA(2000) Cyanobacterial cell walls: News from an unusual prokaryotic envelope. J Bacteriol 182:1191–1199. [4]Wang IN, Smith DL, Young R (2000) Holins: The protein clocks of bacteriophage infections. Annu Rev Microbiol 54:799–825. [5]Garcia-Dominguez M, Lopez-Maury L, Florencio FJ, Reyes JC (2000) A gene clusterinvolved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 182:1507–1514. [6]Warrens AN, Jones MD, Lechler RI (1997) Splicing by overlap extension by PCR using asymmetric amplification: An improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29–35. [7]Akihiro Kato, Kazuhide Use, Nobuyuki Takatani, Kazutaka Ikeda, Miyuki Matsuura, Kouji Kojima (2016) Modulation of the balance of fatty acid production and secretion is crucial for enhancement of growth and productivity of the engineered mutant of the cyanobacterium Synechococcus elongates. Biotechnol Biofuels9:91-101.


