|
|
| (19 intermediate revisions by the same user not shown) |
| Line 117: |
Line 117: |
| | <header> | | <header> |
| | <h2>Harvard BioDesign 2016.</h2> | | <h2>Harvard BioDesign 2016.</h2> |
| − | <p>
| |
| − |
| |
| − | </p>
| |
| | </header> | | </header> |
| | </section> | | </section> |
| | + | |
| | + | |
| | + | |
| | + | <!-- Main --> |
| | + | |
| | + | <div class="wrapper style2"> |
| | + | |
| | + | <article id="main" class="container special"> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <section><center><img src="https://static.igem.org/mediawiki/2016/0/04/T--Harvard_BioDesign--home_logo.png"/></center></section> |
| | + | |
| | + | <header> |
| | + | <h2>Plastiback</h2> |
| | + | <h3>A synthetic biology based plastic sensing feedback device.</h3> |
| | + | <h4>Degrade plastic more simply, efficiently, sustainably. Anywhere.</h4> |
| | + | <h4>Generate electricity from degradation to map out plastic in the ocean.</h4> |
| | + | </header> |
| | + | |
| | + | <section><p><center> |
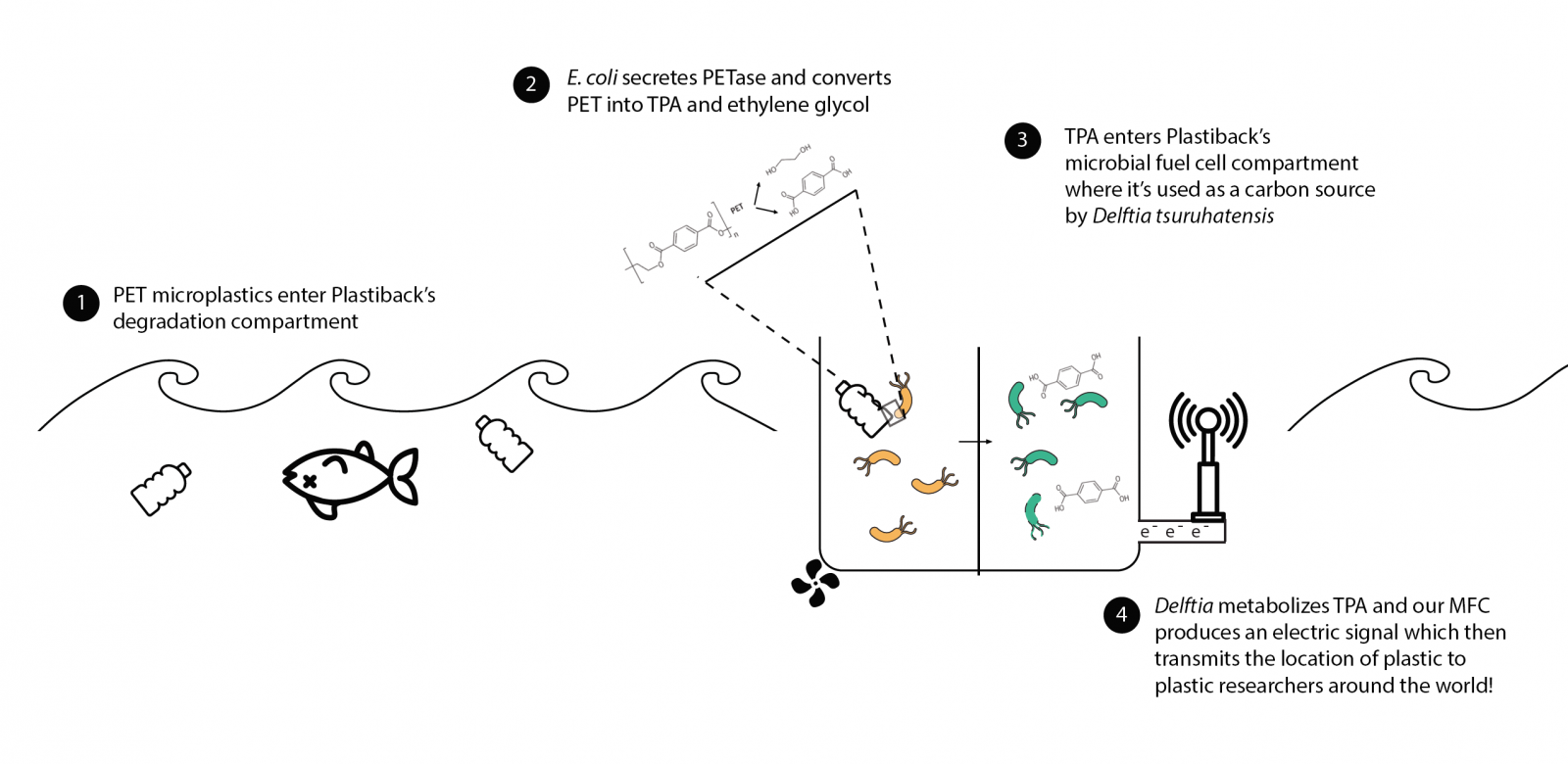
| | + | Plastiback is a plastic-sensing system that gives feedback on where PET, a type of plastic, is located in the ocean. |
| | + | <br>PET is a durable plastic, with multiple uses, including making the bottles for many of the drinks we enjoy. While PET pollutes both the land and sea, the majority of PET litter ends up in the ocean. When plastic enters the ocean, it becomes difficult to clean up and even more difficult to track. For researchers who are investigating the scope and map of where PET is, Plastiback presents a novel and sustainable way to locate ocean PET, while also using non-toxic methods to degrade PET!</center> |
| | + | </p></section> |
| | + | |
| | + | <a href="#" class="image featured"><img src="https://static.igem.org/mediawiki/2016/thumb/d/d6/T--Harvard_BioDesign--home_davidrebekahfinal.png/1600px-T--Harvard_BioDesign--home_davidrebekahfinal.png"/></a> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | </article> |
| | + | |
| | + | </div> |
| | | | |
| | <!-- Carousel --> | | <!-- Carousel --> |
| Line 128: |
Line 160: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Team" class="image featured"><img src="https://static.igem.org/mediawiki/2016/thumb/5/5d/T--Harvard_BioDesign--home_team_3.png/1200px-T--Harvard_BioDesign--home_team_3.png" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Team" class="image featured"><img src="https://static.igem.org/mediawiki/2016/d/d9/T--Harvard_BioDesign--home_team_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Team</h3> | | <h3>Team</h3> |
| Line 135: |
Line 167: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Description" class="image featured"><img src="https://static.igem.org/mediawiki/2016/0/04/T--Harvard_BioDesign--home_logo.png" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Description" class="image featured"><img src="https://static.igem.org/mediawiki/2016/6/6f/T--Harvard_BioDesign--home_project_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Project</h3> | | <h3>Project</h3> |
| Line 142: |
Line 174: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Parts" class="image featured"><img src="images/pic03.jpg" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Parts" class="image featured"><img src="https://static.igem.org/mediawiki/2016/e/e1/T--Harvard_BioDesign--home_parts_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Parts</h3> | | <h3>Parts</h3> |
| Line 150: |
Line 182: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Safety" class="image featured"><img src="images/pic04.jpg" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Safety" class="image featured"><img src="https://static.igem.org/mediawiki/2016/9/94/T--Harvard_BioDesign--home_safety_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Safety</h3> | | <h3>Safety</h3> |
| Line 158: |
Line 190: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Attributions" class="image featured"><img src="images/pic05.jpg" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Attributions" class="image featured"><img src="https://static.igem.org/mediawiki/2016/6/6f/T--Harvard_BioDesign--home_attributions_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Attributions</h3> | | <h3>Attributions</h3> |
| Line 166: |
Line 198: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Human_Practices" class="image featured"><img src="images/pic01.jpg" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Human_Practices" class="image featured"><img src="https://static.igem.org/mediawiki/2016/8/85/T--Harvard_BioDesign--home_human_practices_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Human Practices</h3> | | <h3>Human Practices</h3> |
| Line 174: |
Line 206: |
| | | | |
| | <article> | | <article> |
| − | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Achievements" class="image featured"><img src="images/pic02.jpg" alt="" /></a> | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Achievements" class="image featured"><img src="https://static.igem.org/mediawiki/2016/0/07/T--Harvard_BioDesign--home_achievements_new.png" alt="" /></a> |
| | <header> | | <header> |
| | <h3>Achievements</h3> | | <h3>Achievements</h3> |
| Line 182: |
Line 214: |
| | </div> | | </div> |
| | </section> | | </section> |
| − |
| |
| − | <!-- Main -->
| |
| − |
| |
| − | <div class="wrapper style2">
| |
| − |
| |
| − | <article id="main" class="container special">
| |
| − | <a href="#" class="image featured"><img src="images/pic06.jpg" alt="" /></a>
| |
| − | <header>
| |
| − | <h2>Plastiback</h2>
| |
| − | <p>The Harvard iGEM team began our brainstorming sessions during the late spring. We had a massive brainstorming session where we generated topics to investigate, and in the following weeks, scoured the internet for previous research relating to these topics. We met a few more times to discuss what we had found. After narrowing our search down to 4 projects, we wrote mini proposals to present each idea to the rest of the team. At the beginning of the summer, we continued our project planning with one project: Breaking PET.</p>
| |
| − | <p>In Breaking PET, we hope to engineer a biological system that breaks down PET, a molecule which makes up one sixth of plastic products. Our project is grounded in research done by the Oda group at Kyoto Institute of Technology. In the paper “A bacteria that degrades and assimilates poly(ethylene terephthalate)”, they describe a bacteria found at a bottle recycling plant that has the ability to metabolize PET. From this paper, we found enzyme sequences for the two enzymes thought responsible for PET degradation: PETase and MHETase. PETase breaks down PET into the compound MHET, or mono(2-hydroxylethyl)terephthalic acid. MHETase further breaks down MHET into terephthalic acid and ethylene glycol. The discovery of these two enzymes is exciting because, unlike other PET degrading enzymes, PETase and MHETase are dedicated to the role of PET degradation. Additionally, PETase has been shown to have a degrading efficiency 120 times greater than alternative enzymes.</p>
| |
| − | <p>Our project aims to use E Coli transformed with PETase and MHETase to break down PET and generate electricity. We will create BioBricks for PETase and MHETase and characterize these parts against enzymes used in previous PET degrading iGEM projects. Additionally, we hope to engineer a secretion system for these enzymes. We will also attempt to make a bioreactor/microbial fuel cell to generate electricity from the products of the PET degradation process.</p>
| |
| − |
| |
| − |
| |
| − | </header>
| |
| − | </article>
| |
| − |
| |
| − | </div>
| |
| − |
| |
| − |
| |
| | | | |
| | <!--y Does it start here?--> | | <!--y Does it start here?--> |