|
|
| (43 intermediate revisions by 2 users not shown) |
| Line 1: |
Line 1: |
| − | {{Harvard_BioDesign}}
| + | <!-- |
| | + | Helios by HTML5 UP |
| | + | html5up.net | @ajlkn |
| | + | Free for personal and commercial use under the CCA 3.0 license (html5up.net/license) |
| | + | --> |
| | <html> | | <html> |
| | + | <head> |
| | + | <title>Harvard BioDesign 2016</title> |
| | + | <meta charset="utf-8" /> |
| | + | <meta name="viewport" content="width=device-width, initial-scale=1" /> |
| | | | |
| − | <h3>Project Description</h3> | + | <!--[if lte IE 8]><script src="assets/js/ie/html5shiv.js"></script><![endif]--> |
| − | <p> | + | |
| − | The Harvard iGEM team began our brainstorming sessions during the late spring. We had a massive brainstorming session where we generated topics to investigate, and in the following weeks, scoured the internet for previous research relating to these topics. We met a few more times to discuss what we had found. After narrowing our search down to 4 projects, we wrote mini proposals to present each idea to the rest of the team. At the beginning of the summer, we continued our project planning with one project: Breaking PET.
| + | <link rel="stylesheet" type="text/css" href="https://2016.igem.org/Template:Harvard_BioDesign/assets_css_main?action=raw&ctype=text/css" /> |
| − |
| + | <!--[if lte IE 8]><link rel="stylesheet" href="assets/css/ie8.css" /><![endif]--> |
| − | In Breaking PET, we hope to engineer a biological system that breaks down PET, a molecule which makes up one sixth of plastic products (source). Our project is grounded in research done by the Oda group at Kyoto Institute of Technology. In the paper “A bacteria that degrades and assimilates poly(ethylene terephthalate)”, they describe a bacteria found at a bottle recycling plant that has the ability to metabolize PET. From this paper, we found enzyme sequences for the two enzymes thought responsible for PET degradation: PETase and MHETase. PETase breaks down PET into the compound MHET, or mono(2-hydroxylethyl)terephthalic acid. MHETase further breaks down MHET into terephthalic acid and ethylene glycol. The discovery of these two enzymes is exciting because, unlike other PET degrading enzymes, PETase and MHETase are dedicated to the role of PET degradation. Additionally, PETase has been shown to have a degrading efficiency 120 times greater than alternative enzymes. (source)
| + | |
| | | | |
| − | Our project aims to use E Coli transformed with PETase and MHETase to break down PET and generate electricity. We will create BioBricks for PETase and MHETase and characterize these parts against enzymes used in previous PET degrading iGEM projects. Additionally, we hope to engineer a secretion system for these enzymes. We will also attempt to make a bioreactor/microbial fuel cell to generate electricity from the products of the PET degradation process.
| + | |
| − | </p> | + | <!-- Scripts --> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_jquery_min? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_jquery_dropotron_min? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_jquery_scrolly_min? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_jquery_onvisible_min? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_skel_min? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_util? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | <script type="text/javascript" src="https://2016.igem.org/Template:Harvard_BioDesign/assets_js_main? |
| | + | action=raw&ctype=text/javascript"></script> |
| | + | </head> |
| | + | <body class="homepage"> |
| | + | <div id="page-wrapper"> |
| | | | |
| − | <div class="column full_size" > | + | <!-- Header --> |
| − | <img src="http://placehold.it/800x300/d3d3d3/f2f2f2"> | + | <div id="header_home"> |
| − | </div>
| + | |
| | | | |
| − | <div class="column full_size" > | + | <!-- Inner --> |
| − | <h2> Welcome to iGEM 2016! </h2> | + | <!-- |
| − | <p>Your team has been approved and you are ready to start the iGEM season! </p> | + | <div class="inner"> |
| | + | <header> |
| | + | <h1><a href="https://2016.igem.org/Team:Harvard_BioDesign" id="logo">Harvard BioDesign</a></h1> |
| | + | <hr /> |
| | + | <p>Description</p> |
| | + | </header> |
| | + | <footer> |
| | + | <a href="#banner" class="button circled scrolly">Start</a> |
| | + | </footer> |
| | + | </div> |
| | + | --> |
| | | | |
| − | </div>
| |
| | | | |
| − | <div class="column half_size" > | + | <!-- Nav --> |
| − | <h5>Before you start: </h5> | + | <nav id="nav" > |
| − | <p> Please read the following pages:</p> | + | <ul> |
| − | <ul> | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign">Home</a></li> |
| − | <li> <a href="https://2016.igem.org/Requirements">Requirements page </a> </li> | + | <li> |
| − | <li> <a href="https://2016.igem.org/Wiki_How-To">Wiki Requirements page</a></li> | + | <a href="#">Team</a> |
| − | <li> <a href="https://2016.igem.org/Resources/Template_Documentation"> Template Documentation </a></li> | + | <ul> |
| − | </ul> | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Team">Team</a></li> |
| − | </div> | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Collaborations">Collaborations</a></li> |
| | + | </ul> |
| | + | </li> |
| | + | <li> |
| | + | <a href="#">Project</a> |
| | + | <ul> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Description">Description</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Design">Design</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Experiments">Experiments</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Proof">Proof of Concept</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Demonstrate">Demonstrate</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Results">Results</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Notebook">Notebook</a></li> |
| | + | </ul> |
| | + | </li> |
| | + | <li> |
| | + | <a href="#">Parts</a> |
| | + | <ul> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Parts">Parts</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Basic_Part">Basic Parts</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Composite_Part">Composite Parts</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Part_Collection">Part Collection</a></li> |
| | + | </ul> |
| | + | </li> |
| | | | |
| − | <div class="column half_size" > | + | <!-- R edits --> |
| − | <div class="highlight">
| + | |
| − | <h5> Styling your wiki </h5>
| + | |
| − | <p>You may style this page as you like or you can simply leave the style as it is. You can easily keep the styling and edit the content of these default wiki pages with your project information and completely fulfill the requirement to document your project.</p>
| + | |
| − | <p>While you may not win Best Wiki with this styling, your team is still eligible for all other awards. This default wiki meets the requirements, it improves navigability and ease of use for visitors, and you should not feel it is necessary to style beyond what has been provided.</p>
| + | |
| − | </div>
| + | |
| − | </div>
| + | |
| | | | |
| − | <div class="column full_size" > | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Safety">Safety</a></li> |
| − | <h5> Wiki template information </h5> | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Attributions">Attributions</a></li> |
| − | <p>We have created these wiki template pages to help you get started and to help you think about how your team will be evaluated. You can find a list of all the pages tied to awards here at the <a href="https://2016.igem.org/Judging/Pages_for_Awards/Instructions">Pages for awards</a> link. You must edit these pages to be evaluated for medals and awards, but ultimately the design, layout, style and all other elements of your team wiki is up to you!</p> | + | <li> |
| | + | <a href="#">Human Practices</a> |
| | + | <ul> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Human_Practices">Human Practices</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/HP/Silver">Silver</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/HP/Gold">Gold</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Integrated_Practices">Integrated Practices</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Engagement">Engagement</a></li> |
| | + | </ul> |
| | + | </li> |
| | + | <li> |
| | + | <a href="#">Awards</a> |
| | + | <ul> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Model">Model</a></li> |
| | + | <li><a href="https://2016.igem.org/Team:Harvard_BioDesign/Achievements">Achievements</a></li> |
| | + | </ul> |
| | + | </li> |
| | | | |
| − | </div> | + | </ul> |
| | + | </nav> |
| | | | |
| | + | </div> |
| | | | |
| | + | <!-- Banner --> |
| | + | <section id="banner"> |
| | + | <header> |
| | + | <h2>Harvard BioDesign 2016.</h2> |
| | + | </header> |
| | + | </section> |
| | | | |
| | | | |
| − | <div class="column half_size" >
| |
| − | <h5> Editing your wiki </h5>
| |
| − | <p>On this page you can document your project, introduce your team members, document your progress and share your iGEM experience with the rest of the world! </p>
| |
| − | <p> <a href="https://2016.igem.org/wiki/index.php?title=Team:Example&action=edit"> Click here to edit this page! </a></p>
| |
| | | | |
| − | </div> | + | <!-- Main --> |
| | | | |
| | + | <div class="wrapper style2"> |
| | | | |
| − | <div class="column half_size" > | + | <article id="main" class="container special"> |
| − | <h5>Tips</h5>
| + | |
| − | <p>This wiki will be your team’s first interaction with the rest of the world, so here are a few tips to help you get started: </p>
| + | |
| − | <ul>
| + | |
| − | <li>State your accomplishments! Tell people what you have achieved from the start. </li>
| + | |
| − | <li>Be clear about what you are doing and how you plan to do this.</li>
| + | |
| − | <li>You have a global audience! Consider the different backgrounds that your users come from.</li>
| + | |
| − | <li>Make sure information is easy to find; nothing should be more than 3 clicks away. </li>
| + | |
| − | <li>Avoid using very small fonts and low contrast colors; information should be easy to read. </li>
| + | |
| − | <li>Start documenting your project as early as possible; don’t leave anything to the last minute before the Wiki Freeze. For a complete list of deadlines visit the <a href="https://2016.igem.org/Calendar">iGEM 2016 calendar</a> </li>
| + | |
| − | <li>Have lots of fun! </li>
| + | |
| − | </ul>
| + | |
| − | </div>
| + | |
| | | | |
| | | | |
| − | <div class="column half_size" >
| |
| − | <h5>Inspiration</h5>
| |
| − | <p> You can also view other team wikis for inspiration! Here are some examples:</p>
| |
| − | <ul>
| |
| − | <li> <a href="https://2014.igem.org/Team:SDU-Denmark/"> 2014 SDU Denmark </a> </li>
| |
| − | <li> <a href="https://2014.igem.org/Team:Aalto-Helsinki">2014 Aalto-Helsinki</a> </li>
| |
| − | <li> <a href="https://2014.igem.org/Team:LMU-Munich">2014 LMU-Munich</a> </li>
| |
| − | <li> <a href="https://2014.igem.org/Team:Michigan"> 2014 Michigan</a></li>
| |
| − | <li> <a href="https://2014.igem.org/Team:ITESM-Guadalajara">2014 ITESM-Guadalajara </a></li>
| |
| − | <li> <a href="https://2014.igem.org/Team:SCU-China"> 2014 SCU-China </a></li>
| |
| − | </ul>
| |
| − | </div>
| |
| | | | |
| − | <div class="column half_size" >
| |
| − | <h5> Uploading pictures and files </h5>
| |
| − | <p> You can upload your pictures and files to the iGEM 2016 server. Remember to keep all your pictures and files within your team's namespace or at least include your team's name in the file name. <br />
| |
| − | When you upload, set the "Destination Filename" to <code>Team:YourOfficialTeamName/NameOfFile.jpg</code>. (If you don't do this, someone else might upload a different file with the same "Destination Filename", and your file would be erased!)</p>
| |
| | | | |
| | | | |
| − | <div class="button_click" onClick=" parent.location= 'https://2016.igem.org/Special:Upload '"> | + | <section><center><img src="https://static.igem.org/mediawiki/2016/0/04/T--Harvard_BioDesign--home_logo.png"/></center></section> |
| − | UPLOAD FILES
| + | |
| − | </div> | + | |
| | | | |
| − | </div> | + | <header> |
| | + | <h2>Plastiback</h2> |
| | + | <h3>A synthetic biology based plastic sensing feedback device.</h3> |
| | + | <h4>Degrade plastic more simply, efficiently, sustainably. Anywhere.</h4> |
| | + | <h4>Generate electricity from degradation to map out plastic in the ocean.</h4> |
| | + | </header> |
| | | | |
| | + | <section><p><center> |
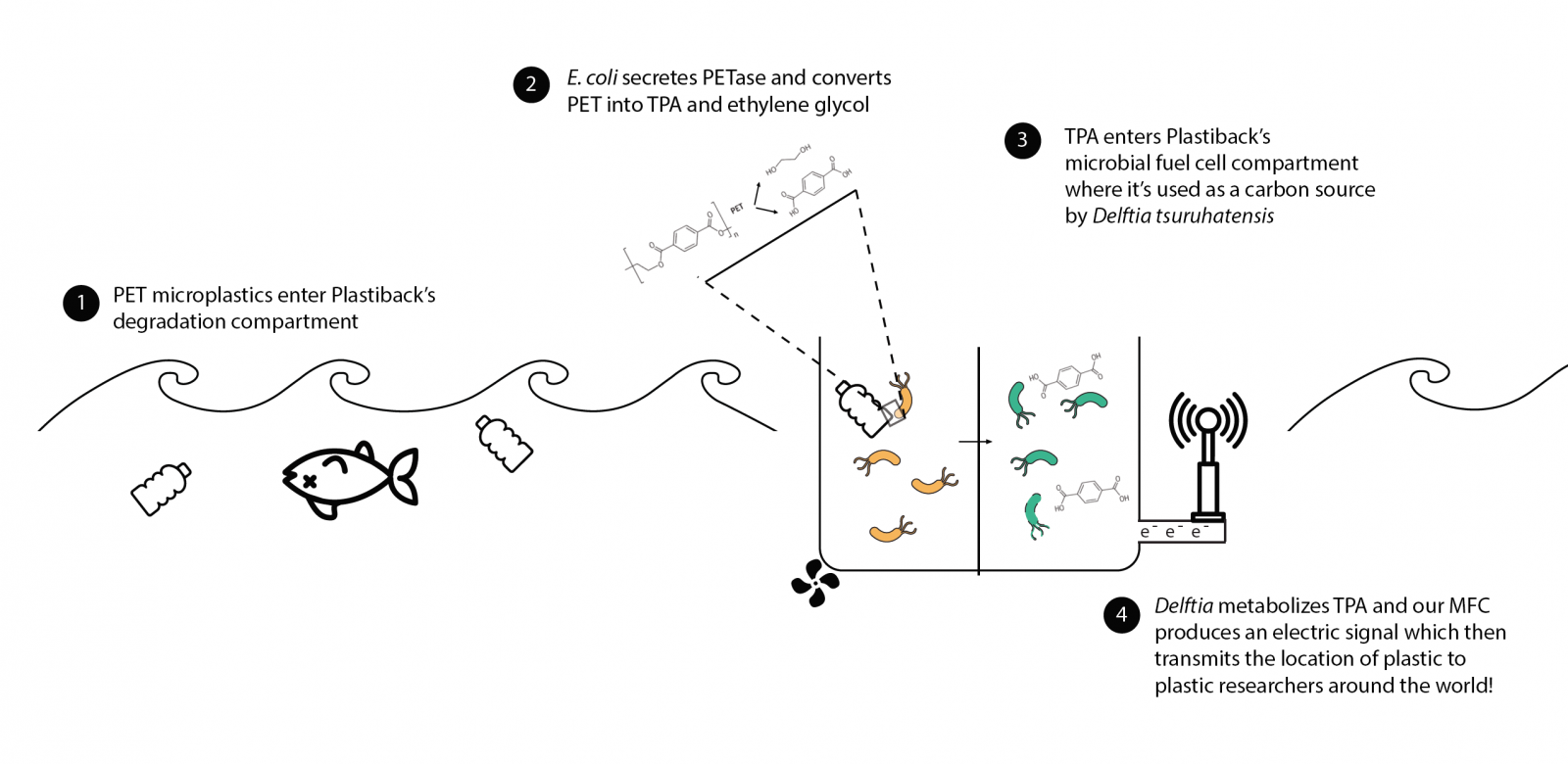
| | + | Plastiback is a plastic-sensing system that gives feedback on where PET, a type of plastic, is located in the ocean. |
| | + | <br>PET is a durable plastic, with multiple uses, including making the bottles for many of the drinks we enjoy. While PET pollutes both the land and sea, the majority of PET litter ends up in the ocean. When plastic enters the ocean, it becomes difficult to clean up and even more difficult to track. For researchers who are investigating the scope and map of where PET is, Plastiback presents a novel and sustainable way to locate ocean PET, while also using non-toxic methods to degrade PET!</center> |
| | + | </p></section> |
| | | | |
| | + | <a href="#" class="image featured"><img src="https://static.igem.org/mediawiki/2016/thumb/d/d6/T--Harvard_BioDesign--home_davidrebekahfinal.png/1600px-T--Harvard_BioDesign--home_davidrebekahfinal.png"/></a> |
| | | | |
| | | | |
| | | | |
| | | | |
| | + | </article> |
| | | | |
| | + | </div> |
| | + | |
| | + | <!-- Carousel --> |
| | + | <section class="carousel"> |
| | + | <div class="reel"> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Team" class="image featured"><img src="https://static.igem.org/mediawiki/2016/d/d9/T--Harvard_BioDesign--home_team_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Team</h3> |
| | + | </header> |
| | + | </article> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Description" class="image featured"><img src="https://static.igem.org/mediawiki/2016/6/6f/T--Harvard_BioDesign--home_project_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Project</h3> |
| | + | </header> |
| | + | </article> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Parts" class="image featured"><img src="https://static.igem.org/mediawiki/2016/e/e1/T--Harvard_BioDesign--home_parts_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Parts</h3> |
| | + | </header> |
| | + | |
| | + | </article> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Safety" class="image featured"><img src="https://static.igem.org/mediawiki/2016/9/94/T--Harvard_BioDesign--home_safety_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Safety</h3> |
| | + | </header> |
| | + | |
| | + | </article> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Attributions" class="image featured"><img src="https://static.igem.org/mediawiki/2016/6/6f/T--Harvard_BioDesign--home_attributions_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Attributions</h3> |
| | + | </header> |
| | + | |
| | + | </article> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Human_Practices" class="image featured"><img src="https://static.igem.org/mediawiki/2016/8/85/T--Harvard_BioDesign--home_human_practices_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Human Practices</h3> |
| | + | </header> |
| | + | |
| | + | </article> |
| | + | |
| | + | <article> |
| | + | <a href="https://2016.igem.org/Team:Harvard_BioDesign/Achievements" class="image featured"><img src="https://static.igem.org/mediawiki/2016/0/07/T--Harvard_BioDesign--home_achievements_new.png" alt="" /></a> |
| | + | <header> |
| | + | <h3>Achievements</h3> |
| | + | </header> |
| | + | </article> |
| | + | |
| | + | </div> |
| | + | </section> |
| | + | |
| | + | <!--y Does it start here?--> |
| | + | <!-- Footer --> |
| | + | <div id="footer"> |
| | + | <div class="container"> |
| | + | <div class="row"> |
| | + | <div class="12u"> |
| | + | |
| | + | <!-- Contact --> |
| | + | <section class="contact"> |
| | + | |
| | + | <ul class="icons"> |
| | + | |
| | + | <li> |
| | + | <a href="https://www.facebook.com/Harvard-iGEM-269794024286" target="_blank"> |
| | + | <img src="https://static.igem.org/mediawiki/2016/2/2f/T--Harvard_BioDesign--images_facebook01.png"alt="Facebook Logo" style="width:51px;height:51px;"> |
| | + | </a> |
| | + | </li> |
| | + | |
| | + | <li> |
| | + | <a href="mailto:crimsonigem@gmail.com"> |
| | + | <img src="https://static.igem.org/mediawiki/2016/e/e2/T--Harvard_BioDesign--images_gmail01.png" alt="Email Logo" style="width:51px;height:51px;"> |
| | + | </a> |
| | + | </li> |
| | + | |
| | + | |
| | + | <li> |
| | + | <a href="https://twitter.com/harvardigem" target="_blank"> |
| | + | <img src="https://static.igem.org/mediawiki/2016/4/4e/T--Harvard_BioDesign--images_twitter01.png"alt="Twitter Logo" style="width:51px;height:51px;"> |
| | + | </a> |
| | + | </li> |
| | + | |
| | + | |
| | + | </ul> |
| | + | </section> |
| | + | |
| | + | <!-- Copyright --> |
| | + | <div class="copyright"> |
| | + | <ul class="menu"> |
| | + | <li>© 2016 Harvard iGEM</li><li>Design: <a href="http://html5up.net">HTML5 UP</a></li> |
| | + | </ul> |
| | + | |
| | + | |
| | + | </div> |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | </div> |
| | + | |
| | + | |
| | + | </body> |
| | </html> | | </html> |