| (12 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
{{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/R-R/style.css}} | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/R-R/style.css}} | ||
{{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Community/css/bootstrap.css}} | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Community/css/bootstrap.css}} | ||
| − | |||
| − | |||
| − | |||
<html> | <html> | ||
| Line 46: | Line 43: | ||
<h1 id="about" class="title text-center">Experiment of <span>CFPS</span></h1> | <h1 id="about" class="title text-center">Experiment of <span>CFPS</span></h1> | ||
<h2><b>Overview</b></h2> | <h2><b>Overview</b></h2> | ||
| − | <p style="font-size:18px">The ability to produce a functional protein in the test tube, rather than in cells, is the essence of cell-free protein synthesis (CFPS)[1].</i> .</p> | + | <p style="font-size:18px">The CFPS system is the assay system in our project for the enzyme modification. The ability to produce a functional protein in the test tube, rather than in cells, is the essence of cell-free protein synthesis (CFPS)[1].</i> .</p> |
<div class="row"> | <div class="row"> | ||
<div class="col-md-2"></div> | <div class="col-md-2"></div> | ||
| Line 63: | Line 60: | ||
</div> | </div> | ||
| − | <p style="font-size:18px">The lack of high-throughput approaches for expression and screening of large enzyme libraries | + | <p style="font-size:18px">The lack of high-throughput approaches for expression and screening of large enzyme libraries is a |
major bottleneck for current enzyme engineering efforts. To address this need, some researchers[2] have developed a high-throughput, fluorescence-based approach for rapid one-pot, microscale expression, and screening of | major bottleneck for current enzyme engineering efforts. To address this need, some researchers[2] have developed a high-throughput, fluorescence-based approach for rapid one-pot, microscale expression, and screening of | ||
| − | different kinds of enzymes. To go further, we try to make our effert to achieve integration of cell-free protein expression with activity screening of enzymes( site-directed | + | different kinds of enzymes. To go further, we try to make our effert to achieve integration of cell-free protein expression with activity screening of enzymes( site-directed mutants of <i>PETase</i>). |
</i> .</p> | </i> .</p> | ||
<div class="row"> | <div class="row"> | ||
| Line 85: | Line 82: | ||
<h2><b>Background</b></h2> | <h2><b>Background</b></h2> | ||
| + | <h3>a.Synthetic Biology</h3> | ||
| + | <p style="font-size:18px">At its core, synthetic biology is inspired by the power and diversity of the living world. What is unique to synthetic biology is the application of an engineering-driven approach to accelerate the design-build-test loops required for reprogramming existing, and constructing new, biological systems. However, a major challenge exists because of our incomplete knowledge of how life works, the daunting complexity of cells, the unintended interference between native and synthetic parts, and (unlike typical engineered systems) the fact that cells evolve, have noise, and have their own agenda such as growth and adaptation[3].In addressing this questions, cell-free systems will play a very inportant role. | ||
| + | |||
| + | </p><br/> | ||
| + | <h3>b.The system</h3> | ||
| + | <p style="font-size:18px">Today cell-free protein synthesis(CFPS) has become a widely used method in molecular biology. In addition to the challenges in vivo above, production of proteins using cell-free protein synthesis usually takes only a few hours, in contrast to production of proteins in cells, which typically takes days to weeks. In fact, even first-time users can often obtain newly synthesized proteins in one day using a commercial system.</p><br/> | ||
| + | |||
| + | <h3>c.Theoretic background</h3> | ||
| + | <p style="font-size:18px">click here to find the<a href="https://2016.igem.org/Team:Tianjin/Protocol">CFPS Protocol</a> | ||
| + | The compounds we will talk about are in the Feeding Buffer of the Protocol, which also plays the role of buffer solution. | ||
| + | For the transcription peorid, from tamplate DNA to mRNA, the necessties including 4 monosomes of RNA, RNA polymersase, energy resourse and other ions. Besides, our CFPS system including the lysate derived from Escherichia coli extract. | ||
| + | The translation machinery is the engine of life. In order to synthesis the protein successfully, the 20 amino acids and ribosomes are obviously necessary. In E.coli CFPS the translation machinery is typically about 20-fold more dilute than in cell, decreasing the rates of initiation, elongation and protein accumulation . As well, the average distance between two adjacent ribosomes on a single mRNA strand increases and polysomes are less likely to form. Despite these differences, CFPS can benefit from the relative slower synthesis rate and the distance between ribosomes by allowing nascent polypeptide chain more time and space to form desirable intra-peptide chain contacts, while decreasing the probability for undesirable, non-specific inter-peptide chain contacts, thereby increasing the probability of proper folding and decreasing the probability of aggregation[4]. As a result, from the CFPS system we can get more enzymes and with higher activity. | ||
| + | </p><br/> | ||
| + | |||
| + | <h3>d.Applications</h3> | ||
<p style="font-size:18px">The diversity of the cell-free systems allows in vitro synthesis of a wide range of proteins for a variety of downstream applications, such as screeening of enzymes activities. In the post-genomic era, cell-free protein synthesis has rapidly become the preferred approach for high-throughput functional and structural studies of proteins and a versatile tool for in vitro protein evolution and synthetic biology.</p> | <p style="font-size:18px">The diversity of the cell-free systems allows in vitro synthesis of a wide range of proteins for a variety of downstream applications, such as screeening of enzymes activities. In the post-genomic era, cell-free protein synthesis has rapidly become the preferred approach for high-throughput functional and structural studies of proteins and a versatile tool for in vitro protein evolution and synthetic biology.</p> | ||
| + | |||
| + | |||
<h2><b>Experiment Design</b></h2> | <h2><b>Experiment Design</b></h2> | ||
| − | <p style="font-size:18px">Basically, we utilized the cell-free system to express the enzymes which had been modified in 22 different sites. We conceived that with this method we could acquire the best modifications by screening them in a high-throughput way. Then we used the proteins we got to degrade PET. </p> | + | |
| + | |||
| + | <p style="font-size:18px">Basically, we utilized the cell-free system to express the enzymes which had been modified in 22 different sites. Besides, we added a fluorescet protein, CFP, before the enzyme. And there is a flexible linker, GGGGSGGGGS , between them. So that we could detect the expression of enzymes by detecting expression of the fluorescent protein with a fluorescence readout instrument, for example, a microplate reader. We conceived that with this method we could acquire the best modifications by screening them in a high-throughput way. Then we used the proteins we got to degrade PET. </p> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | <div class="col-md-8"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/c/ca/T--Tianjin--cf-e3.png" alt="desktop"> | ||
| + | <p style="font-size:15px"> | ||
| + | <br/> | ||
| + | Fig.3.The expression vector in CFPS system | ||
| + | </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
<p style="font-size:18px">How to characterize the degradation velocity is the main problem in our scheme. We analyzed the experiment consequences in two ways. For the first one, we rendered the enzymes degrade pNPa, a general substituent for the detection of PET. Then we measured the absorbance of pNP in the optical density of 400 nanometers, which is the degrading product of pNPa. For the second one, we detected the absorbance of MHET in the optical density of 260 nanometers, which is the product in the first step of PET degradation. | <p style="font-size:18px">How to characterize the degradation velocity is the main problem in our scheme. We analyzed the experiment consequences in two ways. For the first one, we rendered the enzymes degrade pNPa, a general substituent for the detection of PET. Then we measured the absorbance of pNP in the optical density of 400 nanometers, which is the degrading product of pNPa. For the second one, we detected the absorbance of MHET in the optical density of 260 nanometers, which is the product in the first step of PET degradation. | ||
</p> | </p> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | <div class="col-md-8"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/c/c5/T--Tianjin--cf-steps.jpg" alt="desktop"> | ||
| + | <p style="font-size:15px"> | ||
| + | <br/> | ||
| + | Fig.4.Steps for integrated expression and activity screening of enzymes | ||
| + | </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | <h2><b>Reference</b></h2> | ||
| + | |||
| + | <p style="font-size:16px"><i>[1]Gabriel Rosenblum and Barry S. Cooperman. Engine out of the Chassis: Cell-Free Protein Synthesis and its Uses. FEBS Lett. 2014 January 21; 588(2): 261–268. doi:10.1016/j.febslet.2013.10.016.<br/><br/> | ||
| + | [2]Aarthi Chandrasekaran and Anup K. Singh. One-Pot, Microscale Cell-Free Enzyme Expression and Screening. DOI 10.1007/978-1-62703-782-2 Springer New York Heidelberg Dordrecht London.<br/><br/> | ||
| + | [3] C. Eric Hodgman, Michael C. Jewett. Cell-free synthetic biology: Thinking outside the cell. Metabolic Engineering 14 (2012) 261–269. doi:10.1016/j.ymben.2011.09.002<br/><br/> | ||
| + | [4] Shaorong Chong. Overview of Cell-Free Protein Synthesis: Historic Landmarks, Commercial | ||
| + | Systems, and Expanding Applications. Current Protocols in Molecular Biology 16.30.1-16.30.11, October 2014 | ||
| + | DOI: 10.1002/0471142727.mb1630s108 | ||
| + | <br/><br/> | ||
<!-- section end --> | <!-- section end --> | ||
| Line 96: | Line 156: | ||
<!-- section start --> | <!-- section start --> | ||
<!-- ================ --> | <!-- ================ --> | ||
| − | + | </div> | |
| + | </div> | ||
| Line 104: | Line 165: | ||
{{:Team:Tianjin/Templates/Sponsor|}} | {{:Team:Tianjin/Templates/Sponsor|}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 03:49, 20 October 2016
Experiment of CFPS
Overview
The CFPS system is the assay system in our project for the enzyme modification. The ability to produce a functional protein in the test tube, rather than in cells, is the essence of cell-free protein synthesis (CFPS)[1]. .

Fig.1. The basic process of CFPS system
The lack of high-throughput approaches for expression and screening of large enzyme libraries is a major bottleneck for current enzyme engineering efforts. To address this need, some researchers[2] have developed a high-throughput, fluorescence-based approach for rapid one-pot, microscale expression, and screening of different kinds of enzymes. To go further, we try to make our effert to achieve integration of cell-free protein expression with activity screening of enzymes( site-directed mutants of PETase). .

Fig.2.One-pot approach for integrated expression and activity screening of enzymes
Background
a.Synthetic Biology
At its core, synthetic biology is inspired by the power and diversity of the living world. What is unique to synthetic biology is the application of an engineering-driven approach to accelerate the design-build-test loops required for reprogramming existing, and constructing new, biological systems. However, a major challenge exists because of our incomplete knowledge of how life works, the daunting complexity of cells, the unintended interference between native and synthetic parts, and (unlike typical engineered systems) the fact that cells evolve, have noise, and have their own agenda such as growth and adaptation[3].In addressing this questions, cell-free systems will play a very inportant role.
b.The system
Today cell-free protein synthesis(CFPS) has become a widely used method in molecular biology. In addition to the challenges in vivo above, production of proteins using cell-free protein synthesis usually takes only a few hours, in contrast to production of proteins in cells, which typically takes days to weeks. In fact, even first-time users can often obtain newly synthesized proteins in one day using a commercial system.
c.Theoretic background
click here to find theCFPS Protocol The compounds we will talk about are in the Feeding Buffer of the Protocol, which also plays the role of buffer solution. For the transcription peorid, from tamplate DNA to mRNA, the necessties including 4 monosomes of RNA, RNA polymersase, energy resourse and other ions. Besides, our CFPS system including the lysate derived from Escherichia coli extract. The translation machinery is the engine of life. In order to synthesis the protein successfully, the 20 amino acids and ribosomes are obviously necessary. In E.coli CFPS the translation machinery is typically about 20-fold more dilute than in cell, decreasing the rates of initiation, elongation and protein accumulation . As well, the average distance between two adjacent ribosomes on a single mRNA strand increases and polysomes are less likely to form. Despite these differences, CFPS can benefit from the relative slower synthesis rate and the distance between ribosomes by allowing nascent polypeptide chain more time and space to form desirable intra-peptide chain contacts, while decreasing the probability for undesirable, non-specific inter-peptide chain contacts, thereby increasing the probability of proper folding and decreasing the probability of aggregation[4]. As a result, from the CFPS system we can get more enzymes and with higher activity.
d.Applications
The diversity of the cell-free systems allows in vitro synthesis of a wide range of proteins for a variety of downstream applications, such as screeening of enzymes activities. In the post-genomic era, cell-free protein synthesis has rapidly become the preferred approach for high-throughput functional and structural studies of proteins and a versatile tool for in vitro protein evolution and synthetic biology.
Experiment Design
Basically, we utilized the cell-free system to express the enzymes which had been modified in 22 different sites. Besides, we added a fluorescet protein, CFP, before the enzyme. And there is a flexible linker, GGGGSGGGGS , between them. So that we could detect the expression of enzymes by detecting expression of the fluorescent protein with a fluorescence readout instrument, for example, a microplate reader. We conceived that with this method we could acquire the best modifications by screening them in a high-throughput way. Then we used the proteins we got to degrade PET.
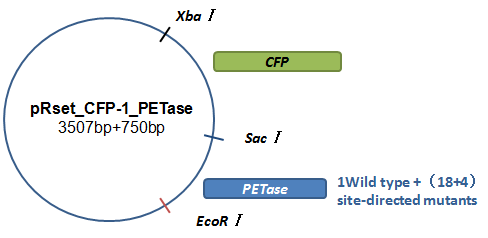
Fig.3.The expression vector in CFPS system
How to characterize the degradation velocity is the main problem in our scheme. We analyzed the experiment consequences in two ways. For the first one, we rendered the enzymes degrade pNPa, a general substituent for the detection of PET. Then we measured the absorbance of pNP in the optical density of 400 nanometers, which is the degrading product of pNPa. For the second one, we detected the absorbance of MHET in the optical density of 260 nanometers, which is the product in the first step of PET degradation.

Fig.4.Steps for integrated expression and activity screening of enzymes
Reference
[1]Gabriel Rosenblum and Barry S. Cooperman. Engine out of the Chassis: Cell-Free Protein Synthesis and its Uses. FEBS Lett. 2014 January 21; 588(2): 261–268. doi:10.1016/j.febslet.2013.10.016.
[2]Aarthi Chandrasekaran and Anup K. Singh. One-Pot, Microscale Cell-Free Enzyme Expression and Screening. DOI 10.1007/978-1-62703-782-2 Springer New York Heidelberg Dordrecht London.
[3] C. Eric Hodgman, Michael C. Jewett. Cell-free synthetic biology: Thinking outside the cell. Metabolic Engineering 14 (2012) 261–269. doi:10.1016/j.ymben.2011.09.002
[4] Shaorong Chong. Overview of Cell-Free Protein Synthesis: Historic Landmarks, Commercial
Systems, and Expanding Applications. Current Protocols in Molecular Biology 16.30.1-16.30.11, October 2014
DOI: 10.1002/0471142727.mb1630s108


