|
|
| (41 intermediate revisions by 5 users not shown) |
| Line 1: |
Line 1: |
| | {{:Team:Tianjin/Templates/MaterialTheme|}} | | {{:Team:Tianjin/Templates/MaterialTheme|}} |
| | | | |
| − | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Note/6803/camarts.css}} | + | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Note/R-R/camarts.css}} |
| | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/css/font.css}} | | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/css/font.css}} |
| − |
| |
| | <html lang="en"> | | <html lang="en"> |
| | + | <style> |
| | + | #Notes{ |
| | + | color: inherit; |
| | + | background-color: rgba(255, 255, 255, 0.1); |
| | + | } |
| | + | </style> |
| | + | |
| | <head> | | <head> |
| | <meta charset="utf-8" /> | | <meta charset="utf-8" /> |
| Line 12: |
Line 18: |
| | | | |
| | <meta content="width=device-width, initial-scale=1.0, maximum-scale=1.0, user-scalable=0" name="viewport" /> | | <meta content="width=device-width, initial-scale=1.0, maximum-scale=1.0, user-scalable=0" name="viewport" /> |
| | + | |
| | + | |
| | + | |
| | + | <style> |
| | + | #Notes{ |
| | + | color: inherit; |
| | + | background-color: rgba(255, 255, 255, 0.1); |
| | + | } |
| | + | </style> |
| | | | |
| | <!------------------------------------------- This part should be dismissed in WIKI u | | <!------------------------------------------- This part should be dismissed in WIKI u |
| Line 43: |
Line 58: |
| | | | |
| | <div id="Topp"></div> | | <div id="Topp"></div> |
| − |
| |
| − |
| |
| | <div id="page" class="hfeed" > | | <div id="page" class="hfeed" > |
| | | | |
| | <div id="page-heading" class="relative"> | | <div id="page-heading" class="relative"> |
| | | | |
| | + | |
| | <header id="main-header" role="banner"> | | <header id="main-header" role="banner"> |
| − | <style>
| + | <style> |
| − | .note-content,.note-content2,.note-content3,.note-content4,.note-content5,.note-content6,.note-content7,.note-content li,.note-content2 li,.note-content3 li,.note-content4 li,.note-content5 li,.note-content6 li,.note-content7 li{ | + | .note-content,.note-content2,.note-content3,.note-content4,.note-content5,.note-content6,.note-content7,.note-content8,.note-content9,.note-content10,.note-content li,.note-content2 li,.note-content3 li,.note-content4 li,.note-content5 li,.note-content6 li,.note-content7 li,.note-content8 li,.note-content9 li,.note-content10 li{ |
| | font-family:Arial; | | font-family:Arial; |
| | font-size:18px; | | font-size:18px; |
| | text-align:justify; | | text-align:justify; |
| | } | | } |
| − | </style> | + | </style> |
| | + | |
| | | | |
| − |
| + | |
| | </div> | | </div> |
| | </header> | | </header> |
| − |
| + | |
| | <div id="main"> | | <div id="main"> |
| | <div id="content" class="home blog single-author one-column content" role="main"> | | <div id="content" class="home blog single-author one-column content" role="main"> |
| − |
| |
| | | | |
| − | <div id="Week1"></div>
| + | <div id="Week1"></div> |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| − | <div class="entry-title" align="center" >Notes For Modified Cyanobacteria:A Controllable Lipid Producer</div> | + | <div class="entry-title" align="center" >Notes For Cyanobacteria</div> |
| | </header><!-- .entry-header --> | | </header><!-- .entry-header --> |
| − | <h1 class="entry-title">Week1(8/1/2016-8/7/2016)</h1>
| |
| − | <div class="entry-content">
| |
| | | | |
| | | | |
| − | <p>
| |
| − | <li>Cultivation: 30µL mother liquid of Synechococcus sp. PCC 7942 (wild type)were cultivated in 10 mL BG-11-media at 30°C and 140rpm.<br/></li>
| |
| − | <li>The genome DNA of 7942 was extracted using a Genomic DNA Isolation Kit.</li>
| |
| − | </p>
| |
| − |
| |
| − |
| |
| − |
| |
| − | <hr class="article">
| |
| − | </article><!-- #post-4252 -->
| |
| | | | |
| − | | + | <!------------------------------------week 1 start------------------------------------------------> |
| − | | + | <div id="Week1"></div> |
| − | | + | |
| − | <div id="Week2"></div>
| + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| − | <h1 class="entry-title">Week2(8/15/2016-8/21/2016)</h1> | + | <h1 class="entry-title">Week1(7/31/2016-8/6/2016)</h1> |
| | </header><!-- .entry-header --> | | </header><!-- .entry-header --> |
| | | | |
| | <div class="entry-content"> | | <div class="entry-content"> |
| − | <p><li>Synechococcus sp PCC 7942 had no signals of life.</li><br />
| + | <div class="note-content2"> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" alt="Igem-6803-week2" width="300px"><br/> | + | |
| − | </a> | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Jul.31th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Cultivation: 30µL mother liquid of Synechococcus sp PCC 7942 (wild type)were cultivated in 10 mL BG-11-media at 30°C and 140rpm.</li> |
| | + | |
| | + | |
| | + | <li>The genome DNA of 7942 was extracted using a Genomic DNA Isolation Kit.</li></div><br/> |
| | + | |
| | + | |
| | + | </div> |
| | + | <div id="Week2"></div> |
| | + | <a class="expand-btn2">Show More</a> |
| | + | |
| | + | |
| | + | |
| | + | </div><!-- .entry-content --> |
| | + | |
| | + | |
| | | | |
| − | <hr class="article">
| |
| | </article> | | </article> |
| | | | |
| | | | |
| | | | |
| − | <div id="Week3"></div>
| + | <!------------------------------------week1 end------------------------------------------------> |
| − | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173">
| + | |
| − | <h1 class="entry-title">Week3(8/29/2016-9/4/2016)</h1> | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <!------------------------------------week2 start------------------------------------------------> |
| | + | |
| | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | + | <h1 class="entry-title">Week2(8/14/2016-8/20/2016)</h1> |
| | <div class="entry-content"> | | <div class="entry-content"> |
| − | <p><li>pMV-G19 and pMV-G15 containing our target genes were received.</li><br/>
| |
| | | | |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" alt="Igem-6803-week3-1" width="300px" ><br/> | + | <div class="note-content4"> |
| − | </a>
| + | |
| | | | |
| − | <li>Colonies were used to inoculate overnight cultures.</li>
| |
| − | <li>Plasmids were isolated using a miniprep kit.</li>
| |
| | | | |
| − | <b>Amplification of <i>19</i> and <i>15</i> with Q5 High-Fidelity DNA Polymerase out of E.coli </b><br/>
| |
| | | | |
| | | | |
| − | <div class="note-content">
| |
| | | | |
| | | | |
| | + | <div style="padding-left:32px;"><li>Synechococcus sp PCC 7942 had no signals of life.</li></div><br/> |
| | | | |
| − | <li><i>19</i> amplification at 65.0°C with 19.rev/fwd primes.</li>
| |
| − | PCR worked, positive control worked, no amplification of <i>19</i>.
| |
| − | <li><i>15</i> amplification at 65.0°C with 15.rev/fwd primes.</li>
| |
| − | PCR worked, positive control worked, no amplification of <i>15</i>.
| |
| − | <li>A product length of 900 bp was expected. The gel shows that most plasmids seem to be correct.</li>
| |
| − | <li>This result was confirmed by sequencing.</li>
| |
| − | <li>The fragments of <i>19</i> and <i>15</i> were purified with PCR Purification Kit.</li>
| |
| | | | |
| − |
| + | |
| − |
| + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" data-lightbox="no" data-title="7942"> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" alt="Igem-6803-week2" width="300px"><br/><br/> |
| | + | </a> |
| | + | <br/> |
| | | | |
| | </div> | | </div> |
| − | <a class="expand-btn">Show More</a>
| |
| | | | |
| | + | <div id="Week3"></div> |
| | + | <a class="expand-btn4">Show More</a> |
| | | | |
| − | <b>Ligation of <i>15</i> with <i>19</i></b><br/>
| |
| | | | |
| − | <div class="note-content2">
| |
| | | | |
| | + | |
| | + | |
| | + | </div><!-- .entry-content --> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | </article> |
| | + | |
| | + | |
| | + | <!------------------------------------week2 end------------------------------------------------> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <!------------------------------------week3 start------------------------------------------------> |
| | + | |
| | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | + | <header class="entry-header"> |
| | + | <h1 class="entry-title">Week3(8/28/2016-9/3/2016)</h1> |
| | + | </header><!-- .entry-header --> |
| | + | |
| | + | <div class="entry-content"> |
| | + | <div class="note-content4"> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Aug.29th</b></h1> |
| | + | <div style="padding-left:32px;"><li>pMV-G19 and pMV-G15 containing our target genes were received.</li><br/> |
| | + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" data-lightbox="no" data-title="pMV-G19 and pMV-G15"> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" alt="Igem-6803-week3-1" width="300px" ><br/><br/> |
| | + | </a> |
| | + | <br/> |
| | + | <li>Colonies were used to inoculate overnight cultures.</li> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Aug.30th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids were isolated using a miniprep kit.</li> |
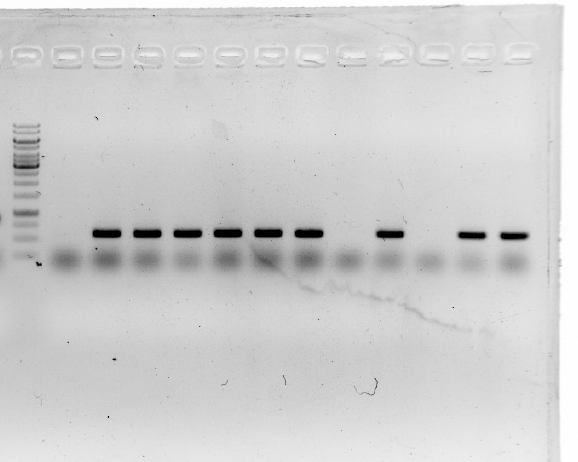
| | + | <li>Amplification of <i>19</i> and <i>15</i> with Q5 High-Fidelity DNA Polymerase out of E.coli.</li> |
| | + | <i>19</i> amplification at 65.0°C with 19.rev/fwd primes.<br/> |
| | + | PCR worked, positive control worked, no amplification of <i>19</i>.<br/> |
| | + | <i>15</i> amplification at 65.0°C with 15.rev/fwd primes.<br/> |
| | + | PCR worked, positive control worked, no amplification of <i>15</i>.<br/> |
| | + | <li>A product length of 900 bp was expected. The gel shows that most plasmids seem to be correct.</li> |
| | + | This result was confirmed by sequencing.<br/> |
| | + | <li>The fragments of <i>19</i> and <i>15</i> were purified with PCR Purification Kit.</li> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Aug.31th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Ligation of <i>15</i> with <i>19</i>.</li> |
| | <li>We add a single 3`-adenine overhang to each end of the fragment of <i>19</i> and then purified with DNA Purification Kit.</li> | | <li>We add a single 3`-adenine overhang to each end of the fragment of <i>19</i> and then purified with DNA Purification Kit.</li> |
| | <li><i>19</i> was ligated into T vectors via TA clone and transformed into E.coli via heat shock.</li> | | <li><i>19</i> was ligated into T vectors via TA clone and transformed into E.coli via heat shock.</li> |
| − | <li>A colony PCR was performed with five colonies.</li> | + | </div> |
| − | Gel electrophoresis showed that 5 colonies were positive for insertion of <i>19</i>.</li>
| + | <br/> |
| − | Two of these colonies containing <i>19</i> were used to inoculate overnight cultures.</li>
| + | |
| | + | <b><h1 style="font-size:108%"> Sep.1th</b></h1> |
| | + | <div style="padding-left:32px;"><li>A colony PCR was performed with five colonies.</li> |
| | + | <li>Gel electrophoresis showed that 5 colonies were positive for insertion of <i>19</i>.</li> |
| | + | <li>Two of these colonies containing <i>19</i> were used to inoculate overnight cultures.</li> |
| | <li><i>15</i> gene fragment was phosphorylated.</li> | | <li><i>15</i> gene fragment was phosphorylated.</li> |
| − | <li>Plasmids containing T vector with <i>19</i>(pT-19)were isolated using a miniprep kit.</li> | + | </div> |
| | + | <br/> |
| | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.2th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids containing T vector with <i>19</i>(pT-19)were isolated using a miniprep kit.</li> |
| | <li>Mono-restriction digest of pT-19 with stu I. </li> | | <li>Mono-restriction digest of pT-19 with stu I. </li> |
| | <li>The enzyme-digested product was dephosphorylation.</li> | | <li>The enzyme-digested product was dephosphorylation.</li> |
| − | <li>Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.</li> | + | <br/> |
| − | Ligation product was transformed into E.coli via heat shock.</li>
| + | |
| − | <li>A colony PCR was performed with twelve colonies.</li> | + | <a href="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" data-lightbox="no" data-title=" "> |
| − | Gel electrophoresis showed that 2 colonies were positive for connecting the plasmid and gene fragment.</li>
| + | |
| − | These two colonies were used to inoculate overnight cultures.</li>
| + | |
| − | <li>Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.</li>
| + | |
| − | <li>Mono-restriction digest of pT-19-15 with Nru I.</li>
| + | |
| − | <li>The enzyme-digested product was dephosphorylation.</li><br/>
| + | |
| | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="800" height="533"><br/> | | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="800" height="533"><br/> |
| | </a> | | </a> |
| − |
| + | <br/> |
| − |
| + | |
| | + | <li>Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.</li> |
| | + | <li>Ligation product was transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| − | <a class="expand-btn2">Show More</a>
| + | <br/> |
| | | | |
| | + | <b><h1 style="font-size:108%"> Sep.3th</b></h1> |
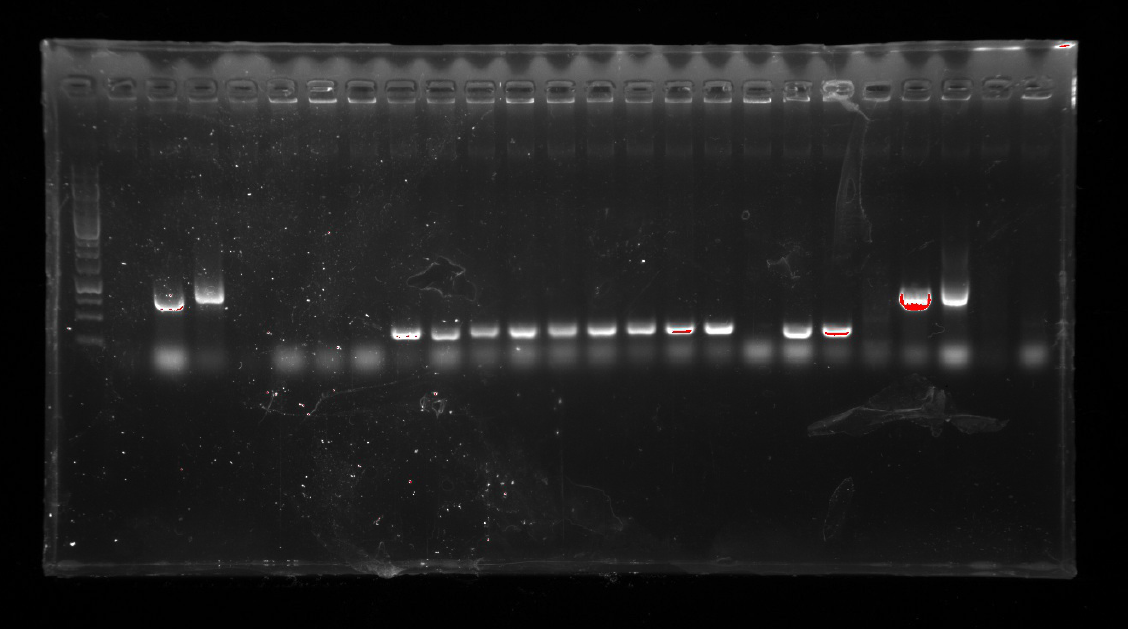
| | + | <div style="padding-left:32px;"><li>A colony PCR was performed with twelve colonies.</li> |
| | + | <li>Gel electrophoresis showed that 2 colonies were positive for connecting the plasmid and gene fragment.</li> |
| | + | <li>These two colonies were used to inoculate overnight cultures.</li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| | | | |
| | + | |
| | + | </div> |
| | + | <div id="Week4"></div> |
| | + | <a class="expand-btn4">Show More</a> |
| | + | |
| | + | |
| | + | </div><!-- .entry-content --> |
| | + | |
| | + | |
| | | | |
| − | <hr class="article">
| |
| | </article> | | </article> |
| | | | |
| | + | <!------------------------------------week3 end------------------------------------------------> |
| | | | |
| | | | |
| − |
| |
| − | <div id="Week4"></div>
| |
| − | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173">
| |
| − | <h1 class="entry-title">Week4(9/5/2016-9/11/2016)</h1>
| |
| − | <div class="entry-content">
| |
| − | <p><li>Colonies containing gene <i>13</i> were used to inoculate overnight cultures.</li>
| |
| − | <li>Plasmids were isolated using a miniprep kit.</li>
| |
| − | <b>Amplification of <i>13</i> with Q5 High-Fidelity DNA Polymerase out of E.coli </b><br/>
| |
| | | | |
| | + | <!------------------------------------week4 start------------------------------------------------> |
| | | | |
| − | <div class="note-content3"> | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | + | <header class="entry-header"> |
| | + | <h1 class="entry-title">Week4(9/4/2016-9/10/2016)</h1> |
| | + | </header><!-- .entry-header --> |
| | | | |
| | + | <div class="entry-content"> |
| | + | <div class="note-content4"> |
| | + | |
| | | | |
| | | | |
| − | <li><i>13</i> amplification at 65.0°C with 13.rev/fwd primes</li>
| |
| − | PCR worked, positive control worked, no amplification of <i>13</i></li>
| |
| − | <li>The fragments of <i>13</i> were purified with PCR Purification Kit.</li>
| |
| − | <li><i>13</i> gene fragment was phosphorylated.</li>
| |
| − |
| |
| − |
| |
| | | | |
| − | </div>
| |
| − | <a class="expand-btn3">Show More</a>
| |
| | | | |
| | + | <b><h1 style="font-size:108%"> Sep.4th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.<li> |
| | + | <li>Mono-restriction digest of pT-19-15 with Nru I.</li> |
| | + | <li>The enzyme-digested product was dephosphorylation.</li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| − | <b>Insertion of Ni promoter and ligation of <i>13-19-15</i></b><br/> | + | <b><h1 style="font-size:108%"> Sep.6th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Colonies containing gene <i>13</i> were used to inoculate overnight cultures. |
| | + | </li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| − | <div class="note-content4"> | + | <b><h1 style="font-size:108%"> Sep.7th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids were isolated using a miniprep kit.</li> |
| | + | <li>Amplification of <i>13</i> with Q5 High-Fidelity DNA Polymerase out of E.coli </li> |
| | + | <li><i>13</i> amplification at 65.0°C with 13.rev/fwd primes</li> |
| | + | PCR worked, positive control worked, no amplification of <i>13</i><br/> |
| | + | <li>The fragments of <i>13</i> were purified with PCR Purification Kit.</li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| | + | <b><h1 style="font-size:108%"> Sep.8th</b></h1> |
| | + | <div style="padding-left:32px;"><li><i>13</i> gene fragment was phosphorylated.</li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.9th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Insertion of Ni promoter and ligation of <i>13-19-15</i></li> |
| | <li>Ni inducible promoter was ligated into pCPC-3301 vector.</li> | | <li>Ni inducible promoter was ligated into pCPC-3301 vector.</li> |
| | <li>Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> | | <li>Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> |
| | <li>Single colonies were obtained by plating.</li> | | <li>Single colonies were obtained by plating.</li> |
| − | <li>A colony PCR of Ni inducible promoter(pCPC-3301-Ni) was performed with 12 colonies.</li> | + | </div> |
| − | Two of the successful ones were used to inoculate overnight cultures.</li> | + | <br/> |
| − | <li>A colony PCR of pT-13-19-15 was performed with 7 colonies.</li> | + | |
| − | Two of the successful ones were used to inoculate overnight cultures.</li>
| + | <b><h1 style="font-size:108%"> Sep.10th</b></h1> |
| − | <li>Two kinds of plasmids were isolated using a miniprep kit.</li><br/>
| + | <div style="padding-left:32px;"> |
| | + | <li>A colony PCR of Ni inducible promoter(pCPC-3301-Ni) was performed with 12 colonies.<li> |
| | + | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| | + | <li>A colony PCR of pT-13-19-15 was performed with 7 colonies.<li> |
| | + | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| | + | <br/> |
| | | | |
| | + | <a href="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" data-lightbox="no" data-title=" "> |
| | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="800" height="533"> <br/> | | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="800" height="533"> <br/> |
| | </a> | | </a> |
| − |
| + | <br/> |
| − |
| + | |
| | </div> | | </div> |
| − | <a class="expand-btn4">Show More</a>
| + | <br/> |
| | | | |
| | + | |
| | + | </div> |
| | + | <div id="Week5"></div> |
| | + | <a class="expand-btn4">Show More</a> |
| | + | |
| | + | </div><!-- .entry-content --> |
| | + | |
| | + | |
| | | | |
| − | <hr class="article"> | + | |
| | </article> | | </article> |
| | | | |
| | + | <!------------------------------------week4 end------------------------------------------------> |
| | | | |
| − | <div id="Week5"></div>
| |
| | | | |
| − | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | + | |
| − | <h1 class="entry-title">Week5(9/12/2016-9/18/2016)</h1> | + | |
| | + | <!------------------------------------week5 start------------------------------------------------> |
| | + | |
| | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | + | <header class="entry-header"> |
| | + | <h1 class="entry-title">Week5(9/11/2016-9/17/2016)</h1> |
| | + | </header><!-- .entry-header --> |
| | + | |
| | <div class="entry-content"> | | <div class="entry-content"> |
| − | <p>
| + | <div class="note-content4"> |
| | | | |
| − | <li>Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.</li> | + | |
| − | <b>The sequencing results for both of them were error.</b> | + | |
| − | <li>Colonies containing Ni inducible promoter were used to inoculate overnight cultures.</li> | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.11th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Two kinds of plasmids were isolated using a miniprep kit. |
| | + | </li> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.14th</b></h1> |
| | + | <div style="padding-left:32px;"><li>The sequencing results for both of them were error.</li> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.15th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Colonies containing Ni inducible promoter were used to inoculate overnight cultures.</li> |
| | <li>PCR was performed to check if the gene fragments were ligated correctly.</li> | | <li>PCR was performed to check if the gene fragments were ligated correctly.</li> |
| − | 13_ fwd and 15_rev on pT-13-19-15<br/>
| + | 13_ fwd and 15_rev on pT-13-19-15<br/> |
| − | Gel electrophoresis showed that it failed.<br/>
| + | <li>Gel electrophoresis showed that it failed.</li> |
| − | <li>Plasmids pCPC-3031-Ni were isolated using a miniprep kit.</li> | + | </div> |
| | + | <br/> |
| | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.16th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids pCPC-3031-Ni were isolated using a miniprep kit.</li> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.17th</b></h1> |
| | + | <div style="padding-left:32px;"> |
| | <li>Several PCRs were performed to check if the gene fragments were ligated correctly.</li> | | <li>Several PCRs were performed to check if the gene fragments were ligated correctly.</li> |
| − | 1.13_fwd and 13_rev on pT-13-19-15<br/>
| + | 13_fwd and 13_rev on pT-13-19-15<br/> |
| − | 2.19_fwd and 19_rev on pT-13-19-15<br/>
| + | 19_fwd and 19_rev on pT-13-19-15<br/> |
| − | 3.15_fwd and 15_rev on pT-13-19-15<br/>
| + | 15_fwd and 15_rev on pT-13-19-15<br/> |
| − | 4.13_fwd and 19_rev on pT-13-19-15<br/>
| + | 13_fwd and 19_rev on pT-13-19-15<br/> |
| − | 5.19_fwd and 15_rev on pT-13-19-15<br/>
| + | 19_fwd and 15_rev on pT-13-19-15<br/> |
| − | 6.13_ wd and 15_rev on pT-13-19-15<br/>
| + | 13_ wd and 15_rev on pT-13-19-15<br/> |
| − | <b>The fourth and sixth ones were not successful.</b><br/>
| + | <b>The fourth and sixth ones were not successful.</b><br/> |
| − | <li><b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> | + | <br/> |
| − | <li><b>Repetition:</b> Several PCRs were performed to check if the gene fragments were ligated correctly.</li>
| + | |
| − | 1.13_fwd and 13_rev on pT-13-19-15<br/>
| + | <a href="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" data-lightbox="no" data-title=" "> |
| − | 2.13_fwd and 19_rev on pT-13-19-15<br/>
| + | |
| − | <b>The second one was failed.</b><br/>
| + | |
| | <img align="center"src="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" alt="Igem-6803-week5" width="800" height="533" ><br/> | | <img align="center"src="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" alt="Igem-6803-week5" width="800" height="533" ><br/> |
| | </a> | | </a> |
| − | | + | <br/> |
| | + | <li><b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> |
| | + | </div> |
| | <br/> | | <br/> |
| | | | |
| Line 263: |
Line 425: |
| | | | |
| | | | |
| | + | </div> |
| | + | <div id="Week6"></div> |
| | + | <a class="expand-btn4">Show More</a> |
| | + | </div><!-- .entry-content --> |
| | + | |
| | + | |
| | | | |
| − | <hr class="article"> | + | |
| | </article> | | </article> |
| | | | |
| | + | <!------------------------------------week5 end------------------------------------------------> |
| | | | |
| | | | |
| | | | |
| | + | <!------------------------------------week6 start------------------------------------------------> |
| | + | |
| | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | + | <header class="entry-header"> |
| | + | <h1 class="entry-title">Week6(9/18/2016-9/24/2016)</h1> |
| | + | </header><!-- .entry-header --> |
| | | | |
| | + | <div class="entry-content"> |
| | | | |
| | + | <div class="note-content4"> |
| | | | |
| | | | |
| | | | |
| | | | |
| − | <div id="Week6"></div>
| |
| − |
| |
| − | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173">
| |
| − | <header class="entry-header">
| |
| − | <h1 class="entry-title">Week6(9/19/2016-9/25/2016)</h1>
| |
| − | </header><!-- .entry-header -->
| |
| | | | |
| − | <div class="entry-content">
| + | <b><h1 style="font-size:108%"> Sep.18th</b></h1> |
| − | <p>
| + | <div style="padding-left:32px;"><b>Repetition:</b> Several PCRs were performed to check if the gene fragments were ligated correctly.<br/> |
| − | <li>Medium preparation :BG-11.</li> | + | 13_fwd and 13_rev on pT-13-19-15<br/> |
| | + | 13_fwd and 19_rev on pT-13-19-15<br/> |
| | + | <b>The second one was failed.</b><br/> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.21th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Medium preparation :BG-11.</li> |
| | <li>19_ fwd and 15_rev were used to amplify <i>19-15</i>.</li> | | <li>19_ fwd and 15_rev were used to amplify <i>19-15</i>.</li> |
| − | Gel electrophoresis showed that amplification of fragments was successfull.</li>
| |
| − | <li>Ligated <i>13</i> and <i>19-15</i> via overlap PCR.</li>
| |
| − | This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.</li>
| |
| − | <li>Restriction digest on pCPC-3031-Ni with Sac I.</li><br/>
| |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"><br/> | + | </div> |
| | + | <br/> |
| | | | |
| − | <li>The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.</li><br/> | + | <b><h1 style="font-size:108%"> Sep.22th</b></h1> |
| | + | <div style="padding-left:32px;"> |
| | + | <li>Gel electrophoresis showed that amplification of fragments was successfull.</li> |
| | + | <li>Ligated <i>13</i> and <i>19-15</i> via overlap PCR.</li> |
| | + | <li>This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.</li> |
| | + | <li>Restriction digest on pCPC-3031-Ni with Sac I.</li> |
| | + | <br/> |
| | + | <a href="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"> |
| | + | </a> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" ><br/> | + | <li>The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.</li> |
| | | | |
| − | <li>A colony PCR of pT-13-19-15 was performed with 12 colonies.</li> | + | <a href="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" data-lightbox="no" data-title=" "> |
| − | Two of these colonies containing pT-13-19-15 were used to inoculate overnight cultures.</li> | + | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" > |
| | + | </a> |
| | + | <br/> |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.23th</b></h1> |
| | + | <div style="padding-left:32px;"><li>A colony PCR of pT-13-19-15 was performed with 12 colonies.</li> |
| | + | <li>Two of these colonies containing pT-13-19-15 were used to inoculate overnight cultures.</li> |
| | <li><i>13-19-15</i> gene fragment was phosphorylated.</li> | | <li><i>13-19-15</i> gene fragment was phosphorylated.</li> |
| | <li>Phosphorylated <i>13-19-15</i> was ligated into pCPC-3031-Ni and transformed into E.coli via heat shock.</li> | | <li>Phosphorylated <i>13-19-15</i> was ligated into pCPC-3031-Ni and transformed into E.coli via heat shock.</li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| − | <li>Plasmids pT-13-19-15 were isolated using a miniprep kit.</li>
| |
| − | <li>A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.</li>
| |
| − | Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.</li><br/>
| |
| | | | |
| | + | <b><h1 style="font-size:108%"> Sep.24th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids pT-13-19-15 were isolated using a miniprep kit.</li> |
| | + | </div> |
| | + | <br/> |
| | | | |
| | + | </div> |
| | + | <div id="Week7"></div> |
| | + | <a class="expand-btn4">Show More</a> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" ><br/>
| |
| − |
| |
| − |
| |
| − | <br />
| |
| | | | |
| | + | </div><!-- .entry-content --> |
| | | | |
| | + | |
| | | | |
| − | <hr class="article"> | + | |
| | </article> | | </article> |
| | | | |
| − | <div id="Week7"></div>
| + | <!------------------------------------week6 end------------------------------------------------> |
| | | | |
| | | | |
| − | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | + | |
| | + | <!------------------------------------week7 start------------------------------------------------> |
| | + | |
| | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| − | <h1 class="entry-title">Week7(9/26/2016-10/2/2016)</h1> | + | <h1 class="entry-title">Week7(9/25/2016-10/1/2016)</h1> |
| | </header><!-- .entry-header --> | | </header><!-- .entry-header --> |
| | | | |
| | <div class="entry-content"> | | <div class="entry-content"> |
| − | <p>
| + | <div class="note-content4"> |
| − | <li>Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.</li> | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.25th</b></h1> |
| | + | <div style="padding-left:32px;"><li>A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.</li> |
| | + | <li>Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.</li> |
| | + | <br/> |
| | + | <a href="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" ><br/> |
| | + | </a> |
| | + | <br/> |
| | + | |
| | + | </div> |
| | + | <br/> |
| | + | |
| | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.26th</b></h1> |
| | + | <div style="padding-left:32px;"><li>Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.</li> |
| | <b>The sequencing results for them were correct.</b> | | <b>The sequencing results for them were correct.</b> |
| | + | <br/> |
| | + | </div> |
| | + | </br> |
| | | | |
| − | <br />
| |
| | | | |
| | | | |
| − | <hr class="article">
| + | <b><h1 style="font-size:108%"> Sep.28th</b></h1> |
| − | </article>
| + | <div style="padding-left:32px;"><li>We transformed the expression vector, pCPC-3031-Ni-13-19-15, into <i>Synechocystis 6803 </i>via electroporation.</li> |
| − | <!-- #post-4252 --> | + | <br/> |
| − |
| + | <b>The story of 6803 is not over. Other team members will show the completed process during the presentation. </br> |
| | + | I will cherish the time spending with you for my whole life. ———by Jiawei Chen and Liangyu Qian</b> |
| | + | </div> |
| | + | |
| | + | |
| | + | |
| | + | </div> |
| | + | <a class="expand-btn4">Show More</a> |
| | + | |
| | + | </div><!-- .entry-content --> |
| | + | |
| | + | |
| | | | |
| | + | </article> |
| | + | |
| | + | <!------------------------------------week7 end------------------------------------------------> |
| | | | |
| | | | |
| | | | |
| − | <!-- #post-4176 -->
| |
| − |
| |
| − |
| |
| | | | |
| | | | |
| Line 358: |
Line 590: |
| | | | |
| | </div> | | </div> |
| − | <div class="side-nav">
| + | |
| − | <ul>
| + | |
| − | <li><a class="topLink" href="#Topp" style="color:#5555FF">Top</a></li>
| + | |
| − | <li><a class="topLink" href="#Week1" style="color:#5555FF">Week1</a></li>
| + | |
| − | <li><a class="topLink" href="#Week2" style="color:#5555FF">Week2</a></li>
| + | |
| − | <li><a class="topLink" href="#Week3" style="color:#5555FF">Week3</a></li>
| + | |
| − | <li><a class="topLink" href="#Week4" style="color:#5555FF">Week4</a></li>
| + | |
| − | <li><a class="topLink" href="#Week5" style="color:#5555FF">Week5</a></li>
| + | |
| − | <li><a class="topLink" href="#Week6" style="color:#5555FF">Week6</a></li>
| + | |
| − | <li><a class="topLink" href="#Week7" style="color:#5555FF">Week7</a></li>
| + | |
| − | </ul>
| + | |
| − | </div>
| + | |
| | | | |
| | | | |
| Line 385: |
Line 606: |
| | </div> | | </div> |
| | | | |
| − | <div id="back-to-top" title="返回本页顶部" >
| + | |
| − | <svg id="rocket" version="1.1" xmlns="http://www.w3.org/2000/svg" width="32" height="32" viewBox="0 0 64 64">
| + | |
| − | <path fill="#B6B6B6" d="M42.057,37.732c0,0,4.139-25.58-9.78-36.207c-0.307-0.233-0.573-0.322-0.802-0.329
| + | |
| − | c-0.227,0.002-0.493,0.083-0.796,0.311c-13.676,10.31-8.95,35.992-8.95,35.992c-10.18,8-7.703,9.151-1.894,23.262
| + | |
| − | c1.108,2.693,3.048,2.06,3.926,0.115c0.877-1.943,2.815-6.232,2.815-6.232l11.029,0.128c0,0,2.035,4.334,2.959,6.298
| + | |
| − | c0.922,1.965,2.877,2.644,3.924-0.024C49.974,47.064,52.423,45.969,42.057,37.732z M31.726,23.155
| + | |
| − | c-2.546-0.03-4.633-2.118-4.664-4.665c-0.029-2.547,2.012-4.587,4.559-4.558c2.546,0.029,4.634,2.117,4.663,4.664
| + | |
| − | C36.314,21.143,34.272,23.184,31.726,23.155z"></path>
| + | |
| − | </svg>
| + | |
| − | </div>
| + | |
| | | | |
| | </body> | | </body> |
| Line 406: |
Line 618: |
| | <!-- Main JS --> | | <!-- Main JS --> |
| | | | |
| − |
| |
| − | <script>
| |
| − | var x = 600;
| |
| − | var y = 2000;
| |
| − | for ( var i = 1; i < 22; i++ ){
| |
| − | var rand = parseInt(Math.random() * (x - y + 1) + y);
| |
| − | var randpic = "http://randomimage.setgetgo.com/get.php?width=" + rand;
| |
| − | $("#MemberName" + i).attr("src", randpic);
| |
| − | }
| |
| − | </script>
| |
| − | <script type="text/javascript">
| |
| − | window._wpemojiSettings = {"baseUrl":"https:\/\/s.w.org\/images\/core\/emoji\/72x72\/","ext":".png","source":{"concatemoji":"http:\/\/www.camarts.cn\/wp-includes\/js\/wp-emoji-release.min.js?ver=4.5.3"}};
| |
| − | !function(a,b,c){function d(a){var c,d,e,f=b.createElement("canvas"),g=f.getContext&&f.getContext("2d"),h=String.fromCharCode;if(!g||!g.fillText)return!1;switch(g.textBaseline="top",g.font="600 32px Arial",a){case"flag":return g.fillText(h(55356,56806,55356,56826),0,0),f.toDataURL().length>3e3;case"diversity":return g.fillText(h(55356,57221),0,0),c=g.getImageData(16,16,1,1).data,d=c[0]+","+c[1]+","+c[2]+","+c[3],g.fillText(h(55356,57221,55356,57343),0,0),c=g.getImageData(16,16,1,1).data,e=c[0]+","+c[1]+","+c[2]+","+c[3],d!==e;case"simple":return g.fillText(h(55357,56835),0,0),0!==g.getImageData(16,16,1,1).data[0];case"unicode8":return g.fillText(h(55356,57135),0,0),0!==g.getImageData(16,16,1,1).data[0]}return!1}function e(a){var c=b.createElement("script");c.src=a,c.type="text/javascript",b.getElementsByTagName("head")[0].appendChild(c)}var f,g,h,i;for(i=Array("simple","flag","unicode8","diversity"),c.supports={everything:!0,everythingExceptFlag:!0},h=0;h<i.length;h++)c.supports[i[h]]=d(i[h]),c.supports.everything=c.supports.everything&&c.supports[i[h]],"flag"!==i[h]&&(c.supports.everythingExceptFlag=c.supports.everythingExceptFlag&&c.supports[i[h]]);c.supports.everythingExceptFlag=c.supports.everythingExceptFlag&&!c.supports.flag,c.DOMReady=!1,c.readyCallback=function(){c.DOMReady=!0},c.supports.everything||(g=function(){c.readyCallback()},b.addEventListener?(b.addEventListener("DOMContentLoaded",g,!1),a.addEventListener("load",g,!1)):(a.attachEvent("onload",g),b.attachEvent("onreadystatechange",function(){"complete"===b.readyState&&c.readyCallback()})),f=c.source||{},f.concatemoji?e(f.concatemoji):f.wpemoji&&f.twemoji&&(e(f.twemoji),e(f.wpemoji)))}(window,document,window._wpemojiSettings);
| |
| − | </script>
| |
| | | | |
| | <script> /* show more */ | | <script> /* show more */ |
| Line 482: |
Line 680: |
| | | | |
| | </script> | | </script> |
| | + | <script> /* show more */ |
| | | | |
| | + | $(document).ready(function() { |
| | + | $( '.expand-btn5' ).click(function() { |
| | + | var $text = $(this).html(); |
| | + | $(this).siblings( '.note-content5' ).toggle(); |
| | + | if( $text === 'Show More' ) { |
| | + | $(this).html( 'Show Less' ); |
| | + | } else { |
| | + | $(this).html( 'Show More' ); |
| | + | } |
| | + | }); |
| | + | }); |
| | | | |
| | + | </script> |
| | + | <script> /* show more */ |
| | + | |
| | + | $(document).ready(function() { |
| | + | $( '.expand-btn6' ).click(function() { |
| | + | var $text = $(this).html(); |
| | + | $(this).siblings( '.note-content6' ).toggle(); |
| | + | if( $text === 'Show More' ) { |
| | + | $(this).html( 'Show Less' ); |
| | + | } else { |
| | + | $(this).html( 'Show More' ); |
| | + | } |
| | + | }); |
| | + | }); |
| | + | |
| | + | </script> |
| | | | |
| | | | |
| Line 504: |
Line 730: |
| | </html> | | </html> |
| | | | |
| − | {{:Team:Tianjin/Templates/AddJS|:Team:Tianjin/Attribution/js/camarts.js}}
| + | |
| | {{:Team:Tianjin/Templates/Sponsor|}} | | {{:Team:Tianjin/Templates/Sponsor|}} |