|
|
| (37 intermediate revisions by 5 users not shown) |
| Line 1: |
Line 1: |
| | {{:Team:Tianjin/Templates/MaterialTheme|}} | | {{:Team:Tianjin/Templates/MaterialTheme|}} |
| | | | |
| − | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Note/Consortium/camarts.css}} | + | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Note/R-R/camarts.css}} |
| | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/css/font.css}} | | {{:Team:Tianjin/Templates/AddCSS|:Team:Tianjin/Experiment/css/font.css}} |
| − |
| |
| | <html lang="en"> | | <html lang="en"> |
| | + | <style> |
| | + | #Notes{ |
| | + | color: inherit; |
| | + | background-color: rgba(255, 255, 255, 0.1); |
| | + | } |
| | + | </style> |
| | + | |
| | <head> | | <head> |
| | <meta charset="utf-8" /> | | <meta charset="utf-8" /> |
| Line 12: |
Line 18: |
| | | | |
| | <meta content="width=device-width, initial-scale=1.0, maximum-scale=1.0, user-scalable=0" name="viewport" /> | | <meta content="width=device-width, initial-scale=1.0, maximum-scale=1.0, user-scalable=0" name="viewport" /> |
| | + | |
| | + | |
| | + | |
| | + | <style> |
| | + | #Notes{ |
| | + | color: inherit; |
| | + | background-color: rgba(255, 255, 255, 0.1); |
| | + | } |
| | + | </style> |
| | | | |
| | <!------------------------------------------- This part should be dismissed in WIKI u | | <!------------------------------------------- This part should be dismissed in WIKI u |
| Line 64: |
Line 79: |
| | <div id="main"> | | <div id="main"> |
| | <div id="content" class="home blog single-author one-column content" role="main"> | | <div id="content" class="home blog single-author one-column content" role="main"> |
| | + | |
| | + | <div id="Week1"></div> |
| | + | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | + | <header class="entry-header"> |
| | + | <div class="entry-title" align="center" >Notes For Cyanobacteria</div> |
| | + | </header><!-- .entry-header --> |
| | | | |
| | | | |
| Line 81: |
Line 102: |
| | | | |
| | <b><h1 style="font-size:108%"> Jul.31th</b></h1> | | <b><h1 style="font-size:108%"> Jul.31th</b></h1> |
| − | <div style="padding-left:32px;">1.Cultivation: 30µL mother liquid of Synechococcus sp PCC 7942 (wild type)were cultivated in 10 mL BG-11-media at 30°C and 140rpm.<br/> | + | <div style="padding-left:32px;"><li>Cultivation: 30µL mother liquid of Synechococcus sp PCC 7942 (wild type)were cultivated in 10 mL BG-11-media at 30°C and 140rpm.</li> |
| − | 2.The genome DNA of 7942 was extracted using a Genomic DNA Isolation Kit.</div><br/>
| + | |
| | + | |
| | + | <li>The genome DNA of 7942 was extracted using a Genomic DNA Isolation Kit.</li></div><br/> |
| | | | |
| | | | |
| | </div> | | </div> |
| | + | <div id="Week2"></div> |
| | <a class="expand-btn2">Show More</a> | | <a class="expand-btn2">Show More</a> |
| | | | |
| Line 107: |
Line 131: |
| | | | |
| | <!------------------------------------week2 start------------------------------------------------> | | <!------------------------------------week2 start------------------------------------------------> |
| − | <div id="Week2"></div>
| + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <h1 class="entry-title">Week2(8/14/2016-8/20/2016)</h1> | | <h1 class="entry-title">Week2(8/14/2016-8/20/2016)</h1> |
| Line 120: |
Line 144: |
| | | | |
| | | | |
| − | <div style="padding-left:32px;">Synechococcus sp PCC 7942 had no signals of life.</div><br/> | + | <div style="padding-left:32px;"><li>Synechococcus sp PCC 7942 had no signals of life.</li></div><br/> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" alt="Igem-6803-week2" width="300px"><br/> | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" data-lightbox="no" data-title="7942"> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/3/37/Igem--6803-week2.jpg" alt="Igem-6803-week2" width="300px"><br/><br/> |
| | </a> | | </a> |
| − |
| + | <br/> |
| | | | |
| | </div> | | </div> |
| | + | |
| | + | <div id="Week3"></div> |
| | <a class="expand-btn4">Show More</a> | | <a class="expand-btn4">Show More</a> |
| | | | |
| Line 151: |
Line 186: |
| | | | |
| | <!------------------------------------week3 start------------------------------------------------> | | <!------------------------------------week3 start------------------------------------------------> |
| − | <div id="Week3"></div> | + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| Line 168: |
Line 203: |
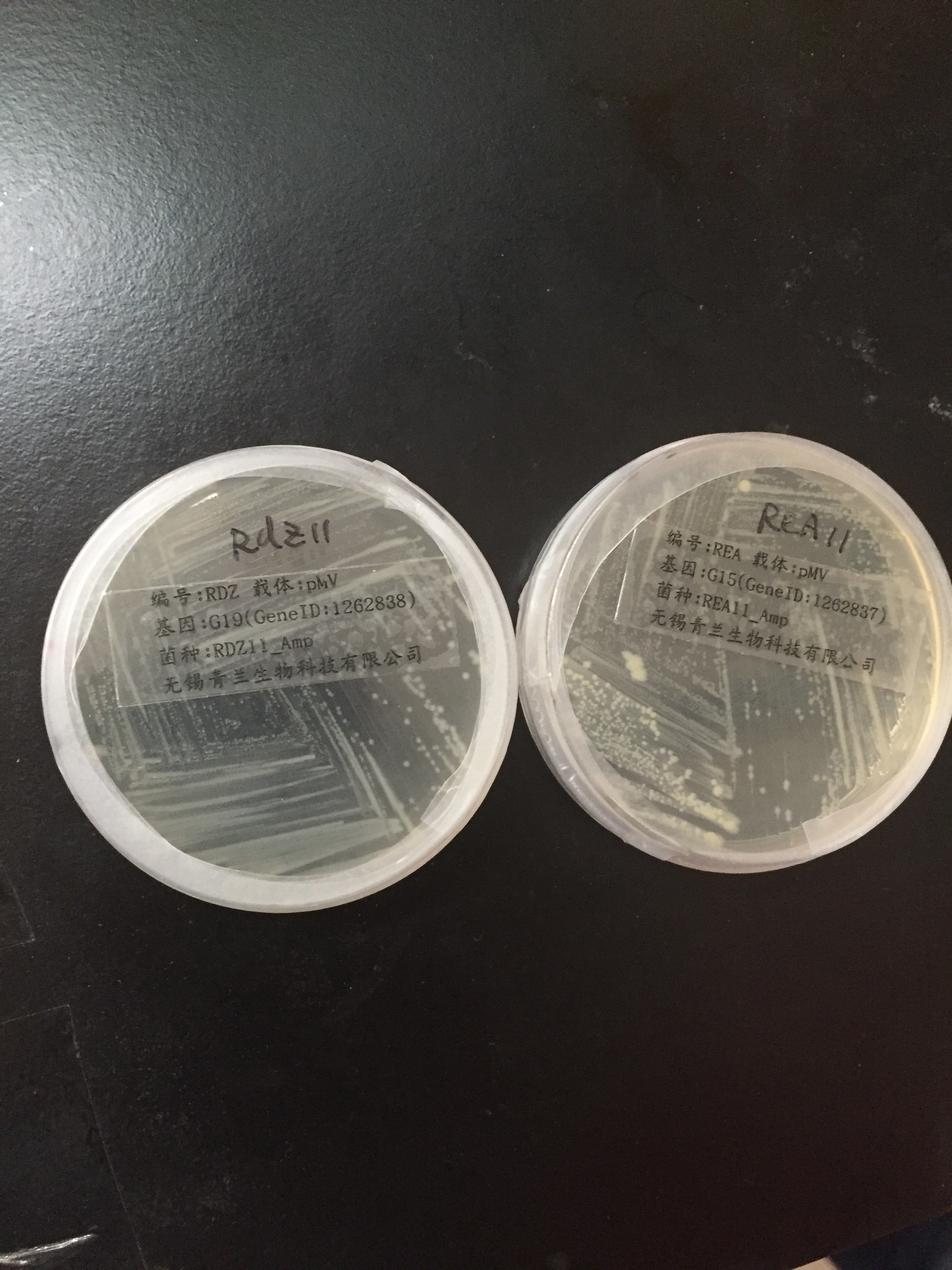
| | <div style="padding-left:32px;"><li>pMV-G19 and pMV-G15 containing our target genes were received.</li><br/> | | <div style="padding-left:32px;"><li>pMV-G19 and pMV-G15 containing our target genes were received.</li><br/> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" alt="Igem-6803-week3-1" width="300px" ><br/> | + | <a href="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" data-lightbox="no" data-title="pMV-G19 and pMV-G15"> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/6/6d/Igem-6803-week3-1.jpg" alt="Igem-6803-week3-1" width="300px" ><br/><br/> |
| | </a> | | </a> |
| − | | + | <br/> |
| − | 2.Colonies were used to inoculate overnight cultures.
| + | <li>Colonies were used to inoculate overnight cultures.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Aug.30th</b></h1> | | <b><h1 style="font-size:108%"> Aug.30th</b></h1> |
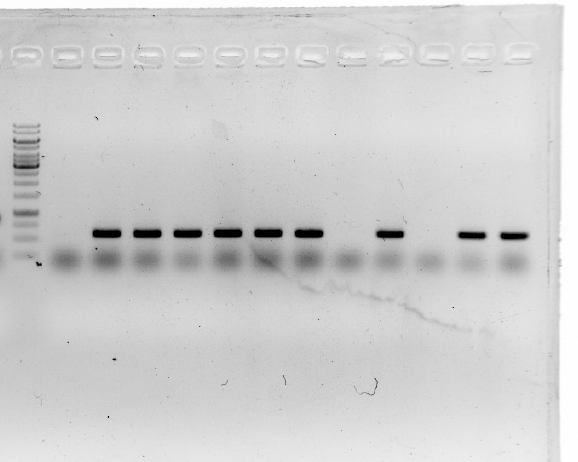
| − | <div style="padding-left:32px;">1.Plasmids were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids were isolated using a miniprep kit.</li> |
| − | 2.Amplification of <i>19</i> and <i>15</i> with Q5 High-Fidelity DNA Polymerase out of E.coli.<br/>
| + | <li>Amplification of <i>19</i> and <i>15</i> with Q5 High-Fidelity DNA Polymerase out of E.coli.</li> |
| | <i>19</i> amplification at 65.0°C with 19.rev/fwd primes.<br/> | | <i>19</i> amplification at 65.0°C with 19.rev/fwd primes.<br/> |
| | PCR worked, positive control worked, no amplification of <i>19</i>.<br/> | | PCR worked, positive control worked, no amplification of <i>19</i>.<br/> |
| | <i>15</i> amplification at 65.0°C with 15.rev/fwd primes.<br/> | | <i>15</i> amplification at 65.0°C with 15.rev/fwd primes.<br/> |
| | PCR worked, positive control worked, no amplification of <i>15</i>.<br/> | | PCR worked, positive control worked, no amplification of <i>15</i>.<br/> |
| − | 3.A product length of 900 bp was expected. The gel shows that most plasmids seem to be correct.<br/>
| + | <li>A product length of 900 bp was expected. The gel shows that most plasmids seem to be correct.</li> |
| | This result was confirmed by sequencing.<br/> | | This result was confirmed by sequencing.<br/> |
| − | 4.The fragments of <i>19</i> and <i>15</i> were purified with PCR Purification Kit.<br/>
| + | <li>The fragments of <i>19</i> and <i>15</i> were purified with PCR Purification Kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Aug.31th</b></h1> | | <b><h1 style="font-size:108%"> Aug.31th</b></h1> |
| − | <div style="padding-left:32px;">1.Ligation of <i>15</i> with <i>19</i>.<br/> | + | <div style="padding-left:32px;"><li>Ligation of <i>15</i> with <i>19</i>.</li> |
| − | 2.We add a single 3`-adenine overhang to each end of the fragment of <i>19</i> and then purified with DNA Purification Kit.<br/>
| + | <li>We add a single 3`-adenine overhang to each end of the fragment of <i>19</i> and then purified with DNA Purification Kit.</li> |
| − | 3.<i>19</i> was ligated into T vectors via TA clone and transformed into E.coli via heat shock.<br/>
| + | <li><i>19</i> was ligated into T vectors via TA clone and transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.1th</b></h1> | | <b><h1 style="font-size:108%"> Sep.1th</b></h1> |
| − | <div style="padding-left:32px;">1.A colony PCR was performed with five colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR was performed with five colonies.</li> |
| − | 2.Gel electrophoresis showed that 5 colonies were positive for insertion of <i>19</i>.<br/>
| + | <li>Gel electrophoresis showed that 5 colonies were positive for insertion of <i>19</i>.</li> |
| − | 3.Two of these colonies containing <i>19</i> were used to inoculate overnight cultures.<br/>
| + | <li>Two of these colonies containing <i>19</i> were used to inoculate overnight cultures.</li> |
| − | 4.<i>15</i> gene fragment was phosphorylated.<br/>
| + | <li><i>15</i> gene fragment was phosphorylated.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 205: |
Line 241: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.2th</b></h1> | | <b><h1 style="font-size:108%"> Sep.2th</b></h1> |
| − | <div style="padding-left:32px;">1.Plasmids containing T vector with <i>19</i>(pT-19)were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids containing T vector with <i>19</i>(pT-19)were isolated using a miniprep kit.</li> |
| − | 2.Mono-restriction digest of pT-19 with stu I. <br/>
| + | <li>Mono-restriction digest of pT-19 with stu I. </li> |
| − | 3.The enzyme-digested product was dephosphorylation.<br/>
| + | <li>The enzyme-digested product was dephosphorylation.</li> |
| − | 4.Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.<br/>
| + | <br/> |
| − | 5.Ligation product was transformed into E.coli via heat shock.<br/>
| + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="800" height="533"><br/> |
| | + | </a> |
| | + | <br/> |
| | + | |
| | + | <li>Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.</li> |
| | + | <li>Ligation product was transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.3th</b></h1> | | <b><h1 style="font-size:108%"> Sep.3th</b></h1> |
| − | <div style="padding-left:32px;">1.A colony PCR was performed with twelve colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR was performed with twelve colonies.</li> |
| − | 2.Gel electrophoresis showed that 2 colonies were positive for connecting the plasmid and gene fragment.<br/>
| + | <li>Gel electrophoresis showed that 2 colonies were positive for connecting the plasmid and gene fragment.</li> |
| − | 3.These two colonies were used to inoculate overnight cultures.<br/>
| + | <li>These two colonies were used to inoculate overnight cultures.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 223: |
Line 266: |
| | | | |
| | </div> | | </div> |
| | + | <div id="Week4"></div> |
| | <a class="expand-btn4">Show More</a> | | <a class="expand-btn4">Show More</a> |
| | | | |
| Line 237: |
Line 281: |
| | | | |
| | <!------------------------------------week4 start------------------------------------------------> | | <!------------------------------------week4 start------------------------------------------------> |
| − | <div id="Week4"></div>
| + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| Line 251: |
Line 295: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.4th</b></h1> | | <b><h1 style="font-size:108%"> Sep.4th</b></h1> |
| − | <div style="padding-left:32px;">1.Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.<li> |
| − | 2.Mono-restriction digest of pT-19-15 with Nru I.<br/>
| + | <li>Mono-restriction digest of pT-19-15 with Nru I.</li> |
| − | 3.The enzyme-digested product was dephosphorylation.<br/>
| + | <li>The enzyme-digested product was dephosphorylation.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.6th</b></h1> | | <b><h1 style="font-size:108%"> Sep.6th</b></h1> |
| − | <div style="padding-left:32px;">Colonies containing gene <i>13</i> were used to inoculate overnight cultures. | + | <div style="padding-left:32px;"><li>Colonies containing gene <i>13</i> were used to inoculate overnight cultures. |
| − | <br/> | + | </li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.7th</b></h1> | | <b><h1 style="font-size:108%"> Sep.7th</b></h1> |
| − | <div style="padding-left:32px;">1.Plasmids were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids were isolated using a miniprep kit.</li> |
| − | 2.Amplification of <i>13</i> with Q5 High-Fidelity DNA Polymerase out of E.coli <br/>
| + | <li>Amplification of <i>13</i> with Q5 High-Fidelity DNA Polymerase out of E.coli </li> |
| − | 3.<i>13</i> amplification at 65.0°C with 13.rev/fwd primes<br/>
| + | <li><i>13</i> amplification at 65.0°C with 13.rev/fwd primes</li> |
| | PCR worked, positive control worked, no amplification of <i>13</i><br/> | | PCR worked, positive control worked, no amplification of <i>13</i><br/> |
| − | 4.The fragments of <i>13</i> were purified with PCR Purification Kit.<br/>
| + | <li>The fragments of <i>13</i> were purified with PCR Purification Kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.8th</b></h1> | | <b><h1 style="font-size:108%"> Sep.8th</b></h1> |
| − | <div style="padding-left:32px;"><i>13</i> gene fragment was phosphorylated.<br/> | + | <div style="padding-left:32px;"><li><i>13</i> gene fragment was phosphorylated.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 279: |
Line 323: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.9th</b></h1> | | <b><h1 style="font-size:108%"> Sep.9th</b></h1> |
| − | <div style="padding-left:32px;">1.Insertion of Ni promoter and ligation of <i>13-19-15</i><br/> | + | <div style="padding-left:32px;"><li>Insertion of Ni promoter and ligation of <i>13-19-15</i></li> |
| − | 2.Ni inducible promoter was ligated into pCPC-3301 vector.<br/>
| + | <li>Ni inducible promoter was ligated into pCPC-3301 vector.</li> |
| − | 3.Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.<br/>
| + | <li>Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> |
| − | 4.Single colonies were obtained by plating.<br/>
| + | <li>Single colonies were obtained by plating.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 288: |
Line 332: |
| | <b><h1 style="font-size:108%"> Sep.10th</b></h1> | | <b><h1 style="font-size:108%"> Sep.10th</b></h1> |
| | <div style="padding-left:32px;"> | | <div style="padding-left:32px;"> |
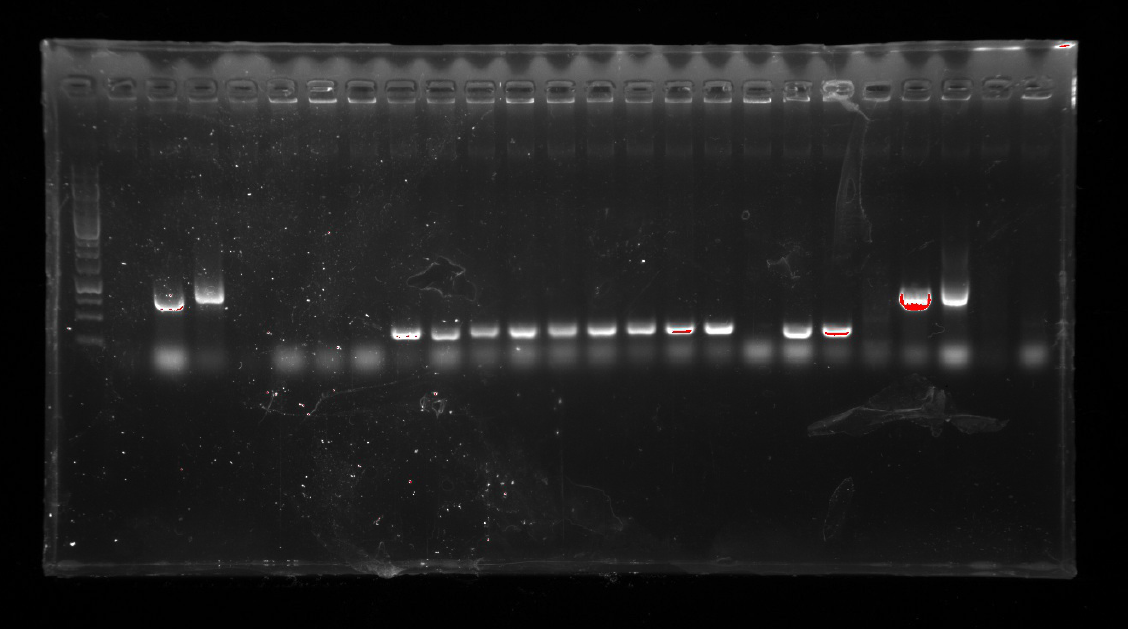
| − | 1.A colony PCR of Ni inducible promoter(pCPC-3301-Ni) was performed with 12 colonies.<br/>
| + | <li>A colony PCR of Ni inducible promoter(pCPC-3301-Ni) was performed with 12 colonies.<li> |
| | Two of the successful ones were used to inoculate overnight cultures.<br/> | | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| − | 2.A colony PCR of pT-13-19-15 was performed with 7 colonies.<br/>
| + | <li>A colony PCR of pT-13-19-15 was performed with 7 colonies.<li> |
| | Two of the successful ones were used to inoculate overnight cultures.<br/> | | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| | + | <br/> |
| | + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="800" height="533"> <br/> |
| | + | </a> |
| | + | <br/> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 297: |
Line 347: |
| | | | |
| | </div> | | </div> |
| | + | <div id="Week5"></div> |
| | <a class="expand-btn4">Show More</a> | | <a class="expand-btn4">Show More</a> |
| | | | |
| Line 312: |
Line 363: |
| | | | |
| | <!------------------------------------week5 start------------------------------------------------> | | <!------------------------------------week5 start------------------------------------------------> |
| − | <div id="Week5"></div>
| + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| Line 320: |
Line 371: |
| | <div class="entry-content"> | | <div class="entry-content"> |
| | <div class="note-content4"> | | <div class="note-content4"> |
| | + | |
| | | | |
| | | | |
| Line 325: |
Line 377: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.11th</b></h1> | | <b><h1 style="font-size:108%"> Sep.11th</b></h1> |
| − | <div style="padding-left:32px;">Two kinds of plasmids were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Two kinds of plasmids were isolated using a miniprep kit. |
| − | </div>
| + | </li> |
| − | <br/>
| + | |
| − | | + | |
| − | <b><h1 style="font-size:108%"> Sep.12th</b></h1>
| + | |
| − | <div style="padding-left:32px;">Two kinds of plasmids were isolated using a miniprep kit. | + | |
| − | <br/> | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.14th</b></h1> | | <b><h1 style="font-size:108%"> Sep.14th</b></h1> |
| − | <div style="padding-left:32px;">The sequencing results for both of them were error.<br/> | + | <div style="padding-left:32px;"><li>The sequencing results for both of them were error.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.15th</b></h1> | | <b><h1 style="font-size:108%"> Sep.15th</b></h1> |
| − | <div style="padding-left:32px;">1.Colonies containing Ni inducible promoter were used to inoculate overnight cultures.<br/> | + | <div style="padding-left:32px;"><li>Colonies containing Ni inducible promoter were used to inoculate overnight cultures.</li> |
| − | 2.PCR was performed to check if the gene fragments were ligated correctly.<br/>
| + | <li>PCR was performed to check if the gene fragments were ligated correctly.</li> |
| | 13_ fwd and 15_rev on pT-13-19-15<br/> | | 13_ fwd and 15_rev on pT-13-19-15<br/> |
| − | 3.Gel electrophoresis showed that it failed.<br/>
| + | <li>Gel electrophoresis showed that it failed.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 350: |
Line 397: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.16th</b></h1> | | <b><h1 style="font-size:108%"> Sep.16th</b></h1> |
| − | <div style="padding-left:32px;">Plasmids pCPC-3031-Ni were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids pCPC-3031-Ni were isolated using a miniprep kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 356: |
Line 403: |
| | <b><h1 style="font-size:108%"> Sep.17th</b></h1> | | <b><h1 style="font-size:108%"> Sep.17th</b></h1> |
| | <div style="padding-left:32px;"> | | <div style="padding-left:32px;"> |
| − | 1.Several PCRs were performed to check if the gene fragments were ligated correctly.<br/>
| + | <li>Several PCRs were performed to check if the gene fragments were ligated correctly.</li> |
| | 13_fwd and 13_rev on pT-13-19-15<br/> | | 13_fwd and 13_rev on pT-13-19-15<br/> |
| | 19_fwd and 19_rev on pT-13-19-15<br/> | | 19_fwd and 19_rev on pT-13-19-15<br/> |
| Line 364: |
Line 411: |
| | 13_ wd and 15_rev on pT-13-19-15<br/> | | 13_ wd and 15_rev on pT-13-19-15<br/> |
| | <b>The fourth and sixth ones were not successful.</b><br/> | | <b>The fourth and sixth ones were not successful.</b><br/> |
| − | 2.<b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.<br/>
| + | <br/> |
| | + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" data-lightbox="no" data-title=" "> |
| | + | <img align="center"src="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" alt="Igem-6803-week5" width="800" height="533" ><br/> |
| | + | </a> |
| | + | <br/> |
| | + | <li><b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 373: |
Line 426: |
| | | | |
| | </div> | | </div> |
| | + | <div id="Week6"></div> |
| | <a class="expand-btn4">Show More</a> | | <a class="expand-btn4">Show More</a> |
| | </div><!-- .entry-content --> | | </div><!-- .entry-content --> |
| Line 386: |
Line 440: |
| | | | |
| | <!------------------------------------week6 start------------------------------------------------> | | <!------------------------------------week6 start------------------------------------------------> |
| − | <div id="Week6"></div> | + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| Line 409: |
Line 463: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.21th</b></h1> | | <b><h1 style="font-size:108%"> Sep.21th</b></h1> |
| − | <div style="padding-left:32px;">1.Medium preparation :BG-11.<br/> | + | <div style="padding-left:32px;"><li>Medium preparation :BG-11.</li> |
| − | 2.19_ fwd and 15_rev were used to amplify <i>19-15</i>.
| + | <li>19_ fwd and 15_rev were used to amplify <i>19-15</i>.</li> |
| − | <br/> | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.22th</b></h1> | | <b><h1 style="font-size:108%"> Sep.22th</b></h1> |
| − | <div style="padding-left:32px;">1.Gel electrophoresis showed that amplification of fragments was successfull.<br/> | + | <div style="padding-left:32px;"> |
| − | 2.Ligated <i>13</i> and <i>19-15</i> via overlap PCR.<br/>
| + | <li>Gel electrophoresis showed that amplification of fragments was successfull.</li> |
| − | 3.This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.<br/>
| + | <li>Ligated <i>13</i> and <i>19-15</i> via overlap PCR.</li> |
| − | 4.Restriction digest on pCPC-3031-Ni with Sac I.<br/>
| + | <li>This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.</li> |
| − | 5.The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.<br/>
| + | <li>Restriction digest on pCPC-3031-Ni with Sac I.</li> |
| | + | <br/> |
| | + | <a href="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"> |
| | + | </a> |
| | + | |
| | + | <li>The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.</li> |
| | + | |
| | + | <a href="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" > |
| | + | </a> |
| | + | <br/> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.23th</b></h1> | | <b><h1 style="font-size:108%"> Sep.23th</b></h1> |
| − | <div style="padding-left:32px;">1.A colony PCR of pT-13-19-15 was performed with 12 colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR of pT-13-19-15 was performed with 12 colonies.</li> |
| − | 2.Two of these colonies containing pT-13-19-15 were used to inoculate overnight cultures.<br/>
| + | <li>Two of these colonies containing pT-13-19-15 were used to inoculate overnight cultures.</li> |
| − | 3.<i>13-19-15</i> gene fragment was phosphorylated.<br/>
| + | <li><i>13-19-15</i> gene fragment was phosphorylated.</li> |
| − | 4.Phosphorylated <i>13-19-15</i> was ligated into pCPC-3031-Ni and transformed into E.coli via heat shock.<br/>
| + | <li>Phosphorylated <i>13-19-15</i> was ligated into pCPC-3031-Ni and transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 434: |
Line 499: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.24th</b></h1> | | <b><h1 style="font-size:108%"> Sep.24th</b></h1> |
| − | <div style="padding-left:32px;">Plasmids pT-13-19-15 were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids pT-13-19-15 were isolated using a miniprep kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | </div> | | </div> |
| | + | <div id="Week7"></div> |
| | <a class="expand-btn4">Show More</a> | | <a class="expand-btn4">Show More</a> |
| | | | |
| Line 454: |
Line 520: |
| | | | |
| | <!------------------------------------week7 start------------------------------------------------> | | <!------------------------------------week7 start------------------------------------------------> |
| − | <div id="Week7"></div>
| + | |
| | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> | | <article id="post-4252" class="post-4252 post type-post status-publish format-standard has-post-thumbnail hentry category-150 tag-174 tag-xinjiang tag-173"> |
| | <header class="entry-header"> | | <header class="entry-header"> |
| Line 468: |
Line 534: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.25th</b></h1> | | <b><h1 style="font-size:108%"> Sep.25th</b></h1> |
| − | <div style="padding-left:32px;">1.A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.</li> |
| − | 2.Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.<br/>
| + | <li>Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.</li> |
| | + | <br/> |
| | + | <a href="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" data-lightbox="no" data-title=" "> |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" ><br/> |
| | + | </a> |
| | + | <br/> |
| | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | + | |
| | + | |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.26th</b></h1> | | <b><h1 style="font-size:108%"> Sep.26th</b></h1> |
| − | <div style="padding-left:32px;">Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.<br/> | + | <div style="padding-left:32px;"><li>Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.</li> |
| | <b>The sequencing results for them were correct.</b> | | <b>The sequencing results for them were correct.</b> |
| | <br/> | | <br/> |
| | </div> | | </div> |
| | + | </br> |
| | + | |
| | + | |
| | + | |
| | + | <b><h1 style="font-size:108%"> Sep.28th</b></h1> |
| | + | <div style="padding-left:32px;"><li>We transformed the expression vector, pCPC-3031-Ni-13-19-15, into <i>Synechocystis 6803 </i>via electroporation.</li> |
| | <br/> | | <br/> |
| | + | <b>The story of 6803 is not over. Other team members will show the completed process during the presentation. </br> |
| | + | I will cherish the time spending with you for my whole life. ———by Jiawei Chen and Liangyu Qian</b> |
| | + | </div> |
| | | | |
| | | | |
| Line 507: |
Line 590: |
| | | | |
| | </div> | | </div> |
| − | <div class="side-nav">
| + | |
| − | <ul>
| + | |
| − | <li><a class="topLink" href="#Topp" style="color:#5555FF">Top</a></li>
| + | |
| − | <li><a class="topLink" href="#Week1" style="color:#5555FF">Week1</a></li>
| + | |
| − | <li><a class="topLink" href="#Week2" style="color:#5555FF">Week2</a></li>
| + | |
| − | <li><a class="topLink" href="#Week3" style="color:#5555FF">Week3</a></li>
| + | |
| − | <li><a class="topLink" href="#Week4" style="color:#5555FF">Week4</a></li>
| + | |
| − | <li><a class="topLink" href="#Week5" style="color:#5555FF">Week5</a></li>
| + | |
| − | <li><a class="topLink" href="#Week6" style="color:#5555FF">Week6</a></li>
| + | |
| − | <li><a class="topLink" href="#Week7" style="color:#5555FF">Week7</a></li>
| + | |
| − |
| + | |
| − | </ul>
| + | |
| − | </div>
| + | |
| | | | |
| | | | |