| Line 154: | Line 154: | ||
$('#minione').animate({"width":"150px"}, 500); | $('#minione').animate({"width":"150px"}, 500); | ||
$('#minitwo').animate({"width":"100px"}, 500); | $('#minitwo').animate({"width":"100px"}, 500); | ||
| − | $('#minithree').animate({"width":"100px"}, 500); | + | $('#minithree').animate({"width":"100px"}, 500); |
| + | $('#minfour').animate({'font-size':'150%'}, 500); | ||
| + | $('#minfour').css('padding', '20px 0px'); | ||
| + | $('#minifour').animate({"width":"100px"}, 500); | ||
$('#hover-contentone').animate({'opacity':'1'}, 1000); | $('#hover-contentone').animate({'opacity':'1'}, 1000); | ||
$('#hover-contenttwo').animate({'opacity':'0'}, 1000); | $('#hover-contenttwo').animate({'opacity':'0'}, 1000); | ||
Revision as of 03:07, 17 October 2016





In the beginning of the summer, we collaborated with MIT, specifically their Recombinase team. Several members of our team worked previously with recombinases and used this time to discuss several topics surrounding this body of work. These topics included structures of recombinase circuits, phylogenetic tree of recombinases, as well as the orthogonality of various heterospecific site-recombinase pairs. In addition, we worked with the entire MIT team to prepare for the NEGEM #1 conference, practicing both presentations and responding to audience questions.
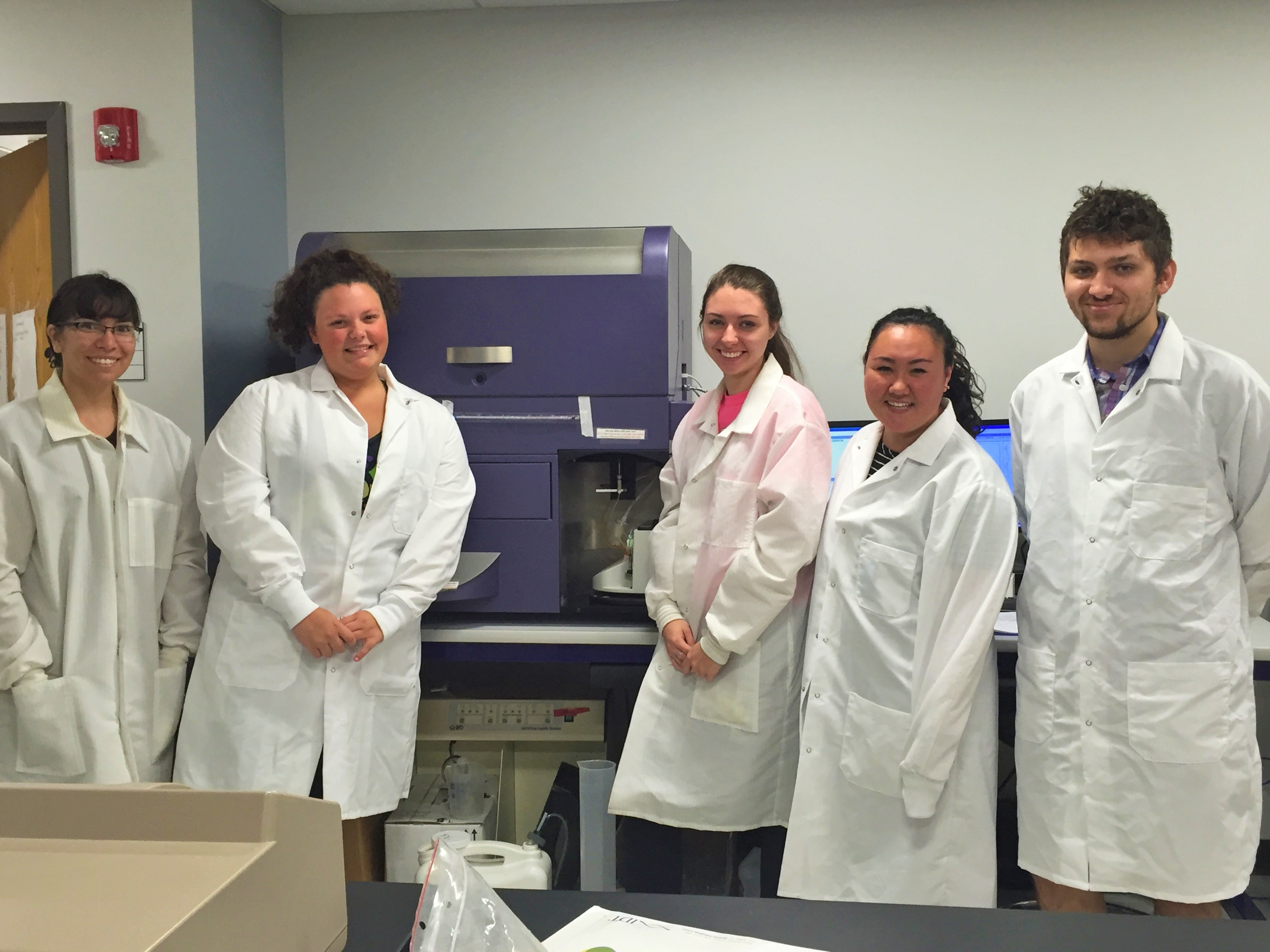
During NEGEM #1, we mingled with the other teams in attendance to potentially collaborate to improve experimental projects or human practices projects. During NEGEM #1 we discussed with WPI how they were using fluorescent microscopy for their assaying technique, while we used flow cytometry. In the true nature of iGEM and further characterization, we proposed a swap: We would run several WPI samples on our flow cytometer and aid in analysis, while WPI would view and image our circuits under their microscopes. In both cases, we were able to validate that our parts functioned as expected. Specifically, we found that our results were replicable under both assaying techniques.

We collaborated with our fellow home team, BostonU HW (Hardware) to develop one of our Human Practices projects, a blog about intellectual property in synthetic biology and software. Uniformly, we agreed that intellectual property was an important topic to iGEMmers and synthetic biologists at large, and thus the disbursement of this information was critical. The first post was a team effort to introduce our readers to intellectual property, patents, copyrights, etc. and to establish the tone for our blogs. Each subsequent post was then switched off so the wet lab team wrote one week and the hardware team wrote the following week. Of course our bloggers, Castor and Pollux, always edited each other's work before it was uploaded.

