| Line 243: | Line 243: | ||
<div class="col-md-3"> | <div class="col-md-3"> | ||
| − | + | <br/><br/> | |
<div align="center"><img src="https://static.igem.org/mediawiki/2016/8/8d/T--Tianjin--experiment-3.png" alt="desktop"></div> | <div align="center"><img src="https://static.igem.org/mediawiki/2016/8/8d/T--Tianjin--experiment-3.png" alt="desktop"></div> | ||
Revision as of 14:25, 11 October 2016
Experiment of Bacteria Consortium
Overview
After Yoshida and his co-workers found and isolated Ideonella sakaiensis 201-F6, which produced two enzymes to degrades PET, we kept very high interests at their works and also came up with many ordinary ideas to increase the efficiency of degradation reaction. Bacteria consortium is one of the most creative ideas.
The inspiration of this idea comes from nature and also learns from nature. Actually, bacteria never exist alone in our nature, they co-work and cooperate together to achieve an aim or live better in a special condition. Thinking from this point, we established a special bacteria consortium for this enzyme catalysis reaction.

Fig.1 Division of work in bacteria consortium
1. Optimization of Culture Conditions
In order to improve efficiency of degrading PET, we are determined to co-culture Pseudomonas putida KT2440, Rhodococcus jostii RHA1 and Bacillus stubtilis 168 (or Bacillus stubtilis DB 104). In our bacteria consortium, work of degradation is divided into several parts as follows:
1.Rhodococcus jostii RHA1 is responsible for degrading TPA (terephthalic acid) to remove substrate inhibition;
2. Pseudomonas putida KT2440 is responsible for degrading EG (ethylene glycol) to remove substrate inhibition, and contribute to produce degradable plastics PHA (polyhydroxyalkanoate);
3. Bacillus stubtilis 168 (or Bacillus stubtilis DB 104) is responsible for secreting PETase and MHETase as the main player of degrading PET.
Whereas, if our bacteria consortium want to achieve this work, they must work in harmony. It is necessary to find an appropriate environment where these bacteria can normally or even better work together.
Primarily, we tried several kinds of media and decide to use W medium as the basic medium in the end; next, we optimized culture conditions by change carbon source, nitrogen source and some ions; then, we checked growing situations and conditions of the degrading PET, TPA and EG; eventually, we could find out the most suitable culture condition to co-culture our bacteria consortium.

Fig.2 Idea about optimization of culture conditions
2. Modification of Pseudomonas putida KT2440
P.putida KT2440 is one of bacteria which can utilize ethylene glycol (EG) at a high speed and meanwhile produces mcl-PHA. In 1988, Lageveen and his co-workers first found mcl-PHA in P.putida KT2440. And then, the metabolism of producing PHA in P.putida KT2440 was reasearched, which found the gene AcoA was the key gene in the procedure. José Manuel Borrero-de Acuña and his co-workers improved the yield by 33% by overexpressing AcoA.
From Björn Mückschel’s works, Ethylene Glycol Metabolism by Pseudomonas putida was found. The key enzymes were identified by comparative proteomics. In P. putida JM37, tartronate semialdehyde synthase (Gcl), malate synthase (GlcB), and isocitrate lyase (AceA) were found to be induced in the presence of ethylene glycol or glyoxylic acid. Under the same conditions, strain KT2440 showed induction of AceA only.
From those studies, we decided to overexpress AcoA and AceA in P.putida KT2440 to help utilize EG as energy source for its growth.
3. Modification of Bacillus subtilis
After some attempt in E.coli and yeast, we look for a new type of host cells- B.subtilis for more secretion. In our experiment, the genes encoding two enzymes are for the first time expressed in S.cerevisiae. Increased yields of PETase and MHETase enzymes are achieved when B. subtilis strains 168 and DB104 (deficient in two and three extracellular proteases, respectively[1]) were transformed with the recombinant plasmid with the help of the enhanced promoter-p43.
4. A Controllable Lipid Producer
Cyanobacteria are excellent organisms for biofuel production. We thus have selected Cyanobacterium Synechocystis sp. PCC 6803 as the source of carbon in our mixed bacteria system. Our target is simply to make the cyanobacteria lyse at the appropriate time by transforming a plasmid contained three bacteriophage-derived lysis genes which were placed downstream of a nickel-inducible signal transduction system into the Synechocystis 6803.
5. Gene Knockout of Escherichia coli
In order to make S. cerevisiae and E.coli better survival, we need to change the energy source from glucose to xylose. When E. coli metabolizes xylose it excretes acetate, which is inhibitory to its own growth. S. cerevisiae cannot metabolize xylose but can use acetate as the sole carbon source without producing ethanol which is toxic to E.coli. For more acetate secretion, we successfully knocked off the atpF and atpH gene from the whole genome with the help of λ-red Recombination and I-SceI Cleavage.
Theoretical Background
1. Degradation of Terephthalate
Rhodococcus sp. strain RHA1 is thought to be capable of degrading a wide range of aromatic compounds including terephthalate acid (TPA). in 2006, a reliable pathway consisting of Distinct ring cleavage dioxygenase systems and protocatechuate (PCA) pathway was come up with, and the proposed degradation pathway for TPA is shown as below[2].

Fig.3 Proposed degradation pathway for TPA[2]

2. Degradation of Ethylene Glycol
By employing growth and bioconversion experiments, directed mutagenesis, and proteome analysis, it is found that Pseudomonas putida KT2440 does not grow within 2 days of incubation, compared to Pseudomonas putida JM37 which can grow rapidly under the same conditions. The key enzymes and specific differences between the two strains were identified by comparative proteomics. In P. putida JM37, tartronate semialdehyde synthase (Gcl), malate synthase (GlcB), and isocitrate lyase (AceA) were found to be induced in the presence of ethylene glycol or glyoxylic acid. Under the same conditions, strain KT2440 showed induction of AceA only. Postulated pathway for the metabolism of ethylene glycol in Pseudomonas putida strains KT2440 and JM37 is shown left[3].
Fig.4 Postulated pathway for the metabolism of ethylene glycol in Pseudomonas putida strains KT2440 and JM37. The enzymes and/or metabolites identified in response to ethylene glycol in KT2440 are depicted in black. Additional pathways identified in strain JM37 are shown in gray. Detailed descriptions of the pathways are given in the text.[3]
3. Production of Polyhydroxyalkanoate
Pseudomonas putida is a natural producer of medium chain length polyhydroxyalkanoates (mcl-PHA), a polymeric precursor of bioplastics. A genome-based in silico model for P. putida KT2440 metabolism was employed to identify potential genetic targets to be engineered for the improvement of mcl-PHA production using glucose as sole carbon source. Here, overproduction of pyruvate dehydrogenase subunit AcoA in the P. putida KT2440 wild type led to an increase of PHA production. In controlled bioreactor batch fermentations PHA production was increased by 33% in the acoA overexpressing wild type in comparison to P. putida KT2440. Transcriptome analyses of engineered PHA producing P. putida in comparison to its parental strains revealed the induction of genes encoding glucose 6-phosphate dehydrogenase and pyruvate dehydrogenase. In addition, NADPH seems to be quantitatively consumed for efficient PHA synthesis, since a direct relationship between low levels of NADPH and high concentrations of the biopolymer were observed. In contrast, intracellular levels of NADH were found increased in PHA producing organisms. Central metabolism of P. putida KT2440 is shown right[4].

Fig.5 Central metabolism of P. putida KT2440[4]
4. Advantages of B. subtilis strains
Bacillus subtilis has an excellent secretion ability, displays fast growth, and is a nonpathogenic bacterium free of endotoxin [5]. It can, therefore, be used in food, enzyme, and pharmaceutical industries and can replace Escherichia coli for protein expression. Furthermore, the extracellular heterogeneous proteins secreted from B. subtilis are more convenient for recovery and purification in large-scale production during downstream processing [6].
5. Enhanced promoter-p43
In order to increase secretion, some enhanced promoters are necessary. However, native gene in a high-copy number plasmid was found to be unstable in B. subtilis[5]. To optimize the production and the stability of the expression vectors, both the promoter and the signal sequence of PETase were replaced by B. subtilis P43 promoter, a constitutively expressed promoter. This overcame the plasmid instability problem.
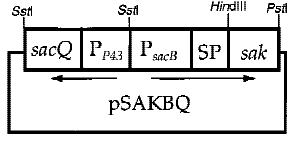
Fig.6 construction of plasmid of cooperation of p43+psacB promoter[7]
6. the cooperation of two promoters-p43+psacB
Interestingly, the cooperation of two promoters in B. subtilis are easily found. For example, An endoglucanase from Bacillus akibai I-1 was successfully overexpressed in Bacillus subtilis 168 by the help of p43 promoter and the expression level of the recombinant enzyme was greatly enhanced by using the sucrose-inducible psacB promoter[7].The construction of plasmid is in the Figure 1 . Thus, we are willing to try whether the combination of p43 and psacB can make a difference in the secretion of enzyme.
Experiment Design
1. Construction of Reporting System
We use a common expression vector plasmid, pUC19, in E.coli to load our device, which consists of heterologous gene part (in this circumstance, PETase gene) and inclusion body reporting part. First of all, we transform the plasmid with part BBa_K339007 from the kit shipped to us using the protocol in the instruction from iGEM official website. Then we use PCR to amplify this part with restriction endonuclease cutting sites Xba1 and Pst1 respectively on sense and anti-sense primers. Then we use corresponding restriction endonuclease to cut the part and plasmid pUC19 and then use T4 DNA ligase to link them together. The next step is to transform the PETase gene into the same plasmid. The initial gene synthetized does not has promoter and terminator so it cannot express. We have to cut the PETase gene and plasmid pET21A with BamH1 and Sal1 enzyme and link them together to transform the PETase gene into pET21A and then use PCR to amplify the T7 promoter-PETase gene-T7 terminator fragment added the restriction endonuclease cutting sites EcoR1 and Sac1. In this way, after we cut the recombinant plasmid pUC19 and T7 promoter-PETase gene-T7 terminator fragment with corresponding restriction endonucleases and link them together, we can obtain the complete device we want.

Fig.8. The construction process of our reporting system
2. Verification of RFP in the part BBa_K339007
The verification of RFP is carried out by using PCR to amplify the RFP gene with restriction endonuclease cutting sites Xba1 and Sac1 added and then cut the RFP and plasmid pET21a with corresponding restriction endonuclease. Then the cut fragments are linked together and transformed into E.coli to express. Then we can detect the red fluorescence.
3. Method of Red Fluorescence Assay
The red fluorescence is detected by 96-well Microplate Reader. The excitation wavelength is set at 584nm and the emission wavelength is set at 607nm. Considering the RFP has an advantage that it can be directly observed by bare eyes, we also use centrifugation to precipitate the bacterial and observe the color of sediment. The red color can be observed if the RFP is expressed. All the experiment including the latter mentioned regulation system use this assay method.
4. Culture and Expression Condition of E.coli in this experiment
Tradition culture medium LB (5g/L yeast extracts, 10g/L peptone, 10g/L NaCl) is also used by us. Because of the ampicillin resistance gene in the plasmid pUC19 and pET21A, ampicillin (100μg/mL) is added to screen for the correctly transformed bacterial. 5mL bacterial are cultured in test tube at 37℃ with 200rpm shaking speed. IPTG is added to induce the expression of PETase gene after 6 hours.

Fig.9. The construction process of our cell lysis based regulation system
5. Construction of Cell Lysis Based Regulation System
This system has a great similarity to the reporting system above. Therefore it is easy to construct because we only need to change the RFP gene to the ddpX gene. However, there is no restriction endonuclease cutting site between the CpxR and RFP gene sequence according to the part map from the iGEM official website, so we have to use PCR to amplify the CpxR promoter solely and add restriction endonuclease cutting sites Xba1 and BamH1 respectively in both end. The ddpX gene is obtained from the E.coli genome using colony PCR and the BamH1 and EcoR1 restriction endonuclease cutting sites are added respectively to both end. Then the three fragments, CpxR promoter, ddpX gene, and cut plasmid pET21a are linked together. Then the whole part is amplified by PCR with Xba1 and Pst1 restriction endonuclease cutting sites added respectively to both end. This way, we can easily cut down the former CpxR-RFP fragment and add the new CpxR-ddpX fragment to the plasmid pUC19.
6. Verification of ddpX Gene Effect
Just like the verification of RFP mentioned before, the verification of ddpX is carried out in the similar way. The pET21a plasmid is cut by BamH1 and EcoR1 instead of Xba1 and EcoR1 , so that the ddpX can be linked to the cut plasmid pET21a solely. Then we can detect if the cell lysis occurs.
7. Method of Cell Lysis Assay
Cell lysis can be reflected by the OD600 of culture medium. The lower the value of OD600 is than the wild type E.coli at the same condition, the stronger the cell lysis effect will be. The OD600 is detected by 96-well Microplate Reader. In order to know the OD600 value continuously, the detection process works through the time of bacterial growth and we will obtain the OD600-Growing time curve.
8. Chassis selection for TPA Positive Feedback Based Regulation System
As the explanation before, the TPA positive feedback system is derived from the TPA degradation metabolic pathway in Rhodococcus jostii RHA1. Considering the difficulty of conducting gene-scale operation in this unusual organism, we directly synthetize all the gene including tpaK, tpaR, and TILS. At first we want to use E.coli to test this device because of the easy and familiar operation. However, in this situation, we have to transform at least 3 plasmids and this cannot be more difficult for E.coli. Therefore, we use another familiar organism, Saccharomyces cerevisiae, as the chassis. In the preliminary experiment, we successfully transform 3 plasmids into Saccharomyces cerevisiae.
9. Construction of TPA Positive Feedback Based Regulation System
We use common plasmids of Saccharomyces cerevisiae, pRS413, pRS415 and pYES2, to respectively load the TPA transporting protein gene, TPA regulation protein gene and TPA induced RFP gene. First of all, we use PCR to amplify all of these fragments and add different restriction endonuclease cutting sites. Then we cut the plasmids with corresponding restriction endonucleases. Then these cut fragments are linked according to the designed order and transformed into Saccharomyces cerevisiae. We screen for the correctly transformed cell by using the Sc-Ura-Leu-His plate.

Fig.10. The construction process of our TPA Positive Feedback Based regulation system
10. Culture and Expression Condition of Saccharomyces cerevisiae in this experiment
Traditional YPD culture medium (22g/L glucose, 20g/L peptone, 10g/L yeast extracts) is used by us. Sc-Ura-Leu-His culture medium (22g/L glucose, 6.7g/L yeast nitrogen base, 1.224g/L nutrient deficiency mixture without Ura, His, Leu and Trp, 5mg/L Trp) is used to screen for correctly transformed cell. All the cells are cultured in 5mL medium at 30℃ with shaking speed of 200rpm. To induce the expression of RFP, we add TPA with different concentration. We first make up TPA standard solution with TPA concentration of 5g/L. Then we respectively add 0, 1μL, 10μL, 100μL, 1mL standard solution to the culture medium.
Expected Results
PETase and MHETase are two key enzymes in our project. However, as heterologous proteins, the expression of these two enzymes face many problems just like expressing other heterologous proteins before including the formation of inclusion body, the lack of regulation pathway, etc. We design this R-R system in order to express the two enzymes visibly and regularly.
First, we hope to directly observe the expression condition of our enzyme by color, when the inclusion body form, which means the overexpressing, the red color can be observed.
Second, when inclusion body form, the normal way to solve this problem is to use lysozyme and ultrasonic to break the cell and purify the protein, which is complex and time-consuming. We expect the cell lysis will automatically occur when the inclusion body form by using ddpX gene.
Third, we expect the chassis organism can sense the existence of TPA, the hydrolyze product of PET and using TPA as the induction of PETase gene. Thus if the degradation process start, this process can be even enhanced until the PET is used up.
References
[1]Physiologie der Mikroorganismen, Humboldt Universitat zu Berlin, Chausseestr. Misfolded maltose binding protein MalE219 induces the CpxRA envelope stress response by stimulating phosphoryl transfer from CpxA to CpxR. Research in Microbiology 160 (2009) 396-400.
[2]Ivan A. D. Lwssard and Christopher T. Walsh. VanX, a bacterial D-alanyl-D-alanine dipeptidase: Resistance, immunity, or survival function? Proc. Natl. Acad. Sci. USA. Vol. 96, pp. 11028–11032, September, 1999.
[3]Hirofumi Hara, Lindsay D. Eltis, Julian E. Davies. Transcriptomic Analysis Reveals a Bifurcated Terephthalate
Degradation Pathway in Rhodococcus sp. Strain RHA1. Journal of Bacteriology, Mar. 2007, 189(5), 1641–1647.
[4]Molina-Henares, A. J., T. Krell, M. E. Guazzaroni, A. Segura, and J. L. Ramos. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30: 157–186.


