|
|
| Line 208: |
Line 208: |
| | 2.Mono-restriction digest of pT-19 with stu I. <br/> | | 2.Mono-restriction digest of pT-19 with stu I. <br/> |
| | 3.The enzyme-digested product was dephosphorylation.<br/> | | 3.The enzyme-digested product was dephosphorylation.<br/> |
| | + | |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="800" height="533"><br/> |
| | + | </a> |
| | + | |
| | + | |
| | 4.Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.<br/> | | 4.Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.<br/> |
| | 5.Ligation product was transformed into E.coli via heat shock.<br/> | | 5.Ligation product was transformed into E.coli via heat shock.<br/> |
| Line 292: |
Line 297: |
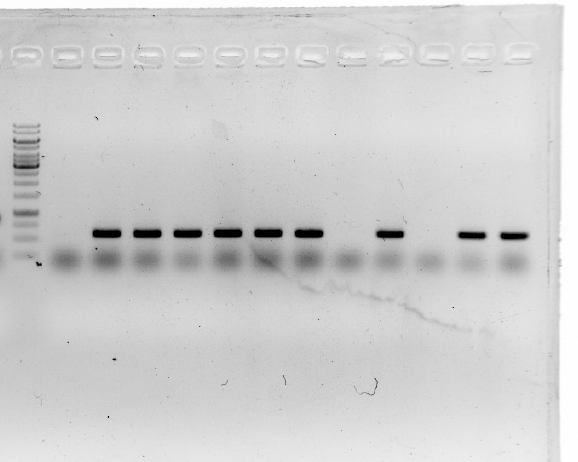
| | 2.A colony PCR of pT-13-19-15 was performed with 7 colonies.<br/> | | 2.A colony PCR of pT-13-19-15 was performed with 7 colonies.<br/> |
| | Two of the successful ones were used to inoculate overnight cultures.<br/> | | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| | + | |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="800" height="533"> <br/> |
| | + | </a> |
| | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 364: |
Line 373: |
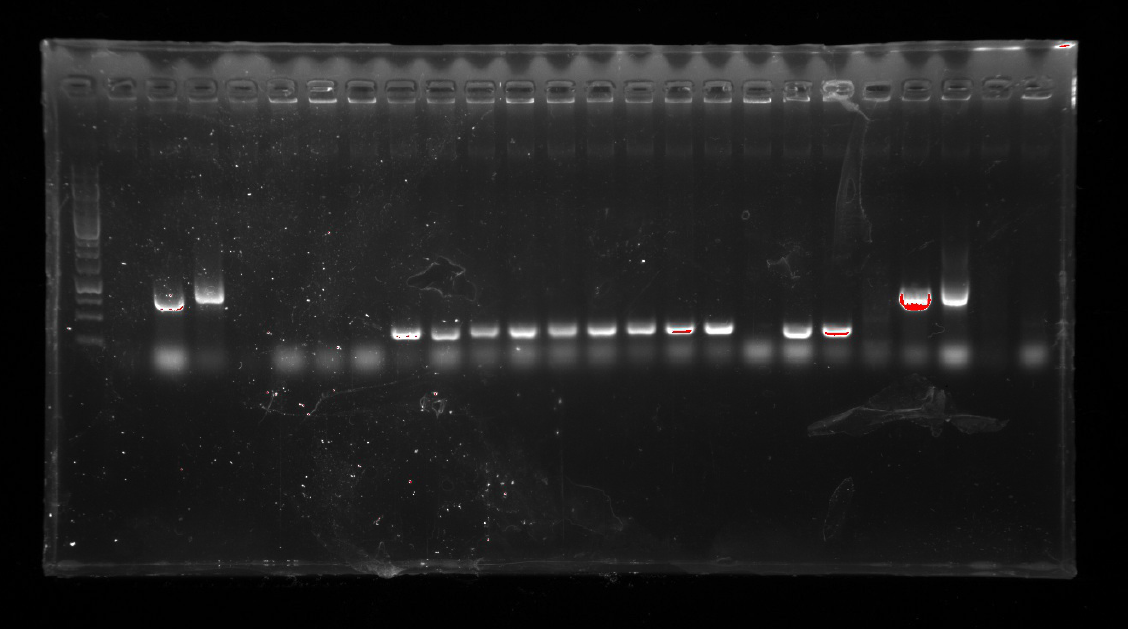
| | 13_ wd and 15_rev on pT-13-19-15<br/> | | 13_ wd and 15_rev on pT-13-19-15<br/> |
| | <b>The fourth and sixth ones were not successful.</b><br/> | | <b>The fourth and sixth ones were not successful.</b><br/> |
| | + | |
| | + | <img align="center"src="https://static.igem.org/mediawiki/2016/4/40/Igem-6803-week5.png" alt="Igem-6803-week5" width="800" height="533" ><br/> |
| | + | </a> |
| | + | |
| | 2.<b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.<br/> | | 2.<b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.<br/> |
| | </div> | | </div> |
| Line 420: |
Line 433: |
| | 3.This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.<br/> | | 3.This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.<br/> |
| | 4.Restriction digest on pCPC-3031-Ni with Sac I.<br/> | | 4.Restriction digest on pCPC-3031-Ni with Sac I.<br/> |
| | + | |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"><br/> |
| | + | |
| | 5.The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.<br/> | | 5.The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.<br/> |
| | + | |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" ><br/> |
| | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 470: |
Line 489: |
| | <div style="padding-left:32px;">1.A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.<br/> | | <div style="padding-left:32px;">1.A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.<br/> |
| | 2.Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.<br/> | | 2.Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.<br/> |
| | + | |
| | + | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" ><br/> |
| | + | |
| | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |