|
|
| Line 203: |
Line 203: |
| | <b><h1 style="font-size:108%"> Sep.1th</b></h1> | | <b><h1 style="font-size:108%"> Sep.1th</b></h1> |
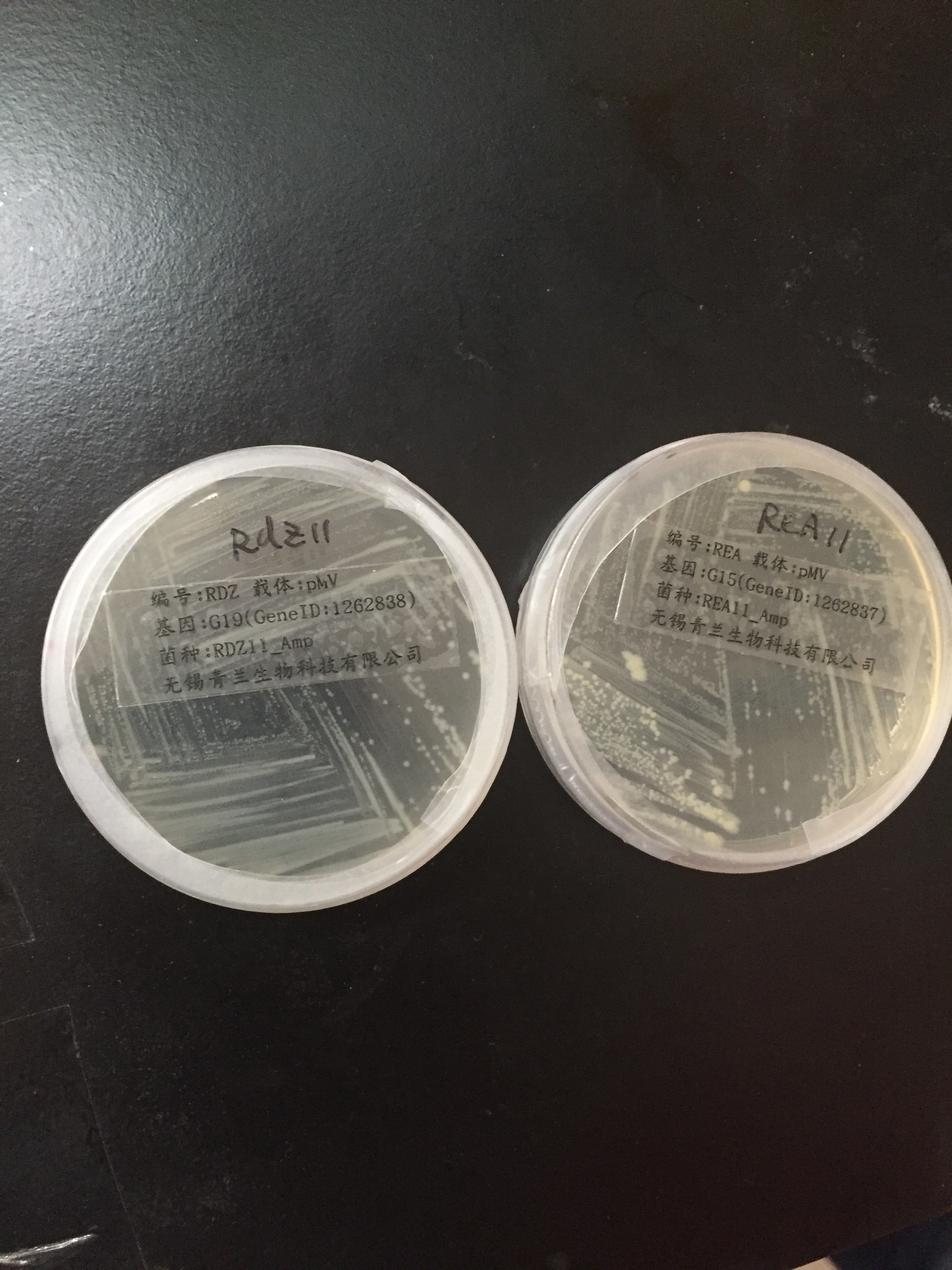
| | <div style="padding-left:32px;"><li>A colony PCR was performed with five colonies.</li> | | <div style="padding-left:32px;"><li>A colony PCR was performed with five colonies.</li> |
| − | 2.Gel electrophoresis showed that 5 colonies were positive for insertion of <i>19</i>.<br/>
| + | <li>Gel electrophoresis showed that 5 colonies were positive for insertion of <i>19</i>.</li> |
| − | 3.Two of these colonies containing <i>19</i> were used to inoculate overnight cultures.<br/>
| + | <li>Two of these colonies containing <i>19</i> were used to inoculate overnight cultures.</li> |
| − | 4.<i>15</i> gene fragment was phosphorylated.<br/>
| + | <li><i>15</i> gene fragment was phosphorylated.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 211: |
Line 211: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.2th</b></h1> | | <b><h1 style="font-size:108%"> Sep.2th</b></h1> |
| − | <div style="padding-left:32px;">1.Plasmids containing T vector with <i>19</i>(pT-19)were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids containing T vector with <i>19</i>(pT-19)were isolated using a miniprep kit.</li> |
| − | 2.Mono-restriction digest of pT-19 with stu I. <br/>
| + | <li>Mono-restriction digest of pT-19 with stu I. </li> |
| − | 3.The enzyme-digested product was dephosphorylation.<br/>
| + | <li>The enzyme-digested product was dephosphorylation.</li> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="60%"><br/> | + | <img align="center" src="https://static.igem.org/mediawiki/2016/a/aa/Igem-6803-week3.png" alt="Igem-6803-week3-2" width="800" height="533"><br/> |
| | </a> | | </a> |
| | | | |
| | | | |
| − | 4.Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.<br/>
| + | <li>Dephosphorylated plasmid and phosphorylated gene <i>15</i> were connected.</li> |
| − | 5.Ligation product was transformed into E.coli via heat shock.<br/>
| + | <li>Ligation product was transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.3th</b></h1> | | <b><h1 style="font-size:108%"> Sep.3th</b></h1> |
| − | <div style="padding-left:32px;">1.A colony PCR was performed with twelve colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR was performed with twelve colonies.</li> |
| − | 2.Gel electrophoresis showed that 2 colonies were positive for connecting the plasmid and gene fragment.<br/>
| + | <li>Gel electrophoresis showed that 2 colonies were positive for connecting the plasmid and gene fragment.</li> |
| − | 3.These two colonies were used to inoculate overnight cultures.<br/>
| + | <li>These two colonies were used to inoculate overnight cultures.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 262: |
Line 262: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.4th</b></h1> | | <b><h1 style="font-size:108%"> Sep.4th</b></h1> |
| − | <div style="padding-left:32px;">1.Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids with correct sequence of <i>19-15</i> were isolated using a miniprep kit.<li> |
| − | 2.Mono-restriction digest of pT-19-15 with Nru I.<br/>
| + | <li>Mono-restriction digest of pT-19-15 with Nru I.</li> |
| − | 3.The enzyme-digested product was dephosphorylation.<br/>
| + | <li>The enzyme-digested product was dephosphorylation.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 275: |
Line 275: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.7th</b></h1> | | <b><h1 style="font-size:108%"> Sep.7th</b></h1> |
| − | <div style="padding-left:32px;">1.Plasmids were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids were isolated using a miniprep kit.</li> |
| − | 2.Amplification of <i>13</i> with Q5 High-Fidelity DNA Polymerase out of E.coli <br/>
| + | <li>Amplification of <i>13</i> with Q5 High-Fidelity DNA Polymerase out of E.coli </li> |
| − | 3.<i>13</i> amplification at 65.0°C with 13.rev/fwd primes<br/>
| + | <li><i>13</i> amplification at 65.0°C with 13.rev/fwd primes</li> |
| | PCR worked, positive control worked, no amplification of <i>13</i><br/> | | PCR worked, positive control worked, no amplification of <i>13</i><br/> |
| − | 4.The fragments of <i>13</i> were purified with PCR Purification Kit.<br/>
| + | <li>The fragments of <i>13</i> were purified with PCR Purification Kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 290: |
Line 290: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.9th</b></h1> | | <b><h1 style="font-size:108%"> Sep.9th</b></h1> |
| − | <div style="padding-left:32px;">1.Insertion of Ni promoter and ligation of <i>13-19-15</i><br/> | + | <div style="padding-left:32px;"><li>Insertion of Ni promoter and ligation of <i>13-19-15</i></li> |
| − | 2.Ni inducible promoter was ligated into pCPC-3301 vector.<br/>
| + | <li>Ni inducible promoter was ligated into pCPC-3301 vector.</li> |
| − | 3.Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.<br/>
| + | <li>Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> |
| − | 4.Single colonies were obtained by plating.<br/>
| + | <li>Single colonies were obtained by plating.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 299: |
Line 299: |
| | <b><h1 style="font-size:108%"> Sep.10th</b></h1> | | <b><h1 style="font-size:108%"> Sep.10th</b></h1> |
| | <div style="padding-left:32px;"> | | <div style="padding-left:32px;"> |
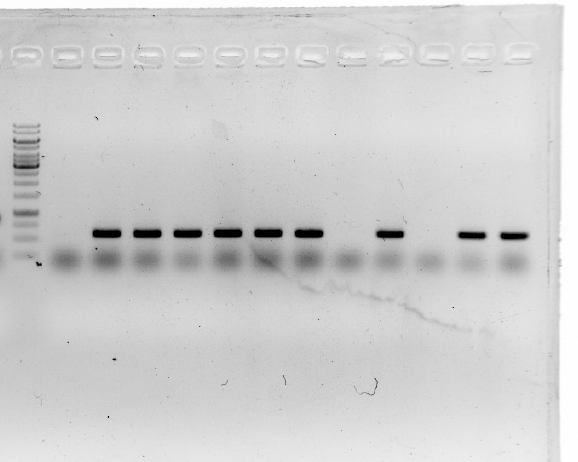
| − | 1.A colony PCR of Ni inducible promoter(pCPC-3301-Ni) was performed with 12 colonies.<br/>
| + | <li>A colony PCR of Ni inducible promoter(pCPC-3301-Ni) was performed with 12 colonies.<li> |
| | Two of the successful ones were used to inoculate overnight cultures.<br/> | | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| − | 2.A colony PCR of pT-13-19-15 was performed with 7 colonies.<br/>
| + | <li>A colony PCR of pT-13-19-15 was performed with 7 colonies.<li> |
| | Two of the successful ones were used to inoculate overnight cultures.<br/> | | Two of the successful ones were used to inoculate overnight cultures.<br/> |
| | | | |
| − | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="500px"> <br/> | + | <img align="center" src="https://static.igem.org/mediawiki/2016/8/81/Igem-6803-week4.png" alt="Igem-6803-week4" width="800" height="533"> <br/> |
| | </a> | | </a> |
| | | | |
| Line 356: |
Line 356: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.15th</b></h1> | | <b><h1 style="font-size:108%"> Sep.15th</b></h1> |
| − | <div style="padding-left:32px;">1.Colonies containing Ni inducible promoter were used to inoculate overnight cultures.<br/> | + | <div style="padding-left:32px;"><li>Colonies containing Ni inducible promoter were used to inoculate overnight cultures.</li> |
| − | 2.PCR was performed to check if the gene fragments were ligated correctly.<br/>
| + | <li>PCR was performed to check if the gene fragments were ligated correctly.</li> |
| | 13_ fwd and 15_rev on pT-13-19-15<br/> | | 13_ fwd and 15_rev on pT-13-19-15<br/> |
| − | 3.Gel electrophoresis showed that it failed.<br/>
| + | <li>Gel electrophoresis showed that it failed.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 365: |
Line 365: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.16th</b></h1> | | <b><h1 style="font-size:108%"> Sep.16th</b></h1> |
| − | <div style="padding-left:32px;">Plasmids pCPC-3031-Ni were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids pCPC-3031-Ni were isolated using a miniprep kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 371: |
Line 371: |
| | <b><h1 style="font-size:108%"> Sep.17th</b></h1> | | <b><h1 style="font-size:108%"> Sep.17th</b></h1> |
| | <div style="padding-left:32px;"> | | <div style="padding-left:32px;"> |
| − | 1.Several PCRs were performed to check if the gene fragments were ligated correctly.<br/>
| + | <li>Several PCRs were performed to check if the gene fragments were ligated correctly.</li> |
| | 13_fwd and 13_rev on pT-13-19-15<br/> | | 13_fwd and 13_rev on pT-13-19-15<br/> |
| | 19_fwd and 19_rev on pT-13-19-15<br/> | | 19_fwd and 19_rev on pT-13-19-15<br/> |
| Line 383: |
Line 383: |
| | </a> | | </a> |
| | | | |
| − | 2.<b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.<br/>
| + | <li><b>Repetition:</b> Phosphorylated <i>13</i> was ligated into pT-19-15 and transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 428: |
Line 428: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.21th</b></h1> | | <b><h1 style="font-size:108%"> Sep.21th</b></h1> |
| − | <div style="padding-left:32px;">1.Medium preparation :BG-11.<br/> | + | <div style="padding-left:32px;"><li>Medium preparation :BG-11.</li> |
| − | 2.19_ fwd and 15_rev were used to amplify <i>19-15</i>.
| + | <li>19_ fwd and 15_rev were used to amplify <i>19-15</i>.</li> |
| − | <br/> | + | |
| | </div> | | </div> |
| | <br/> | | <br/> |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.22th</b></h1> | | <b><h1 style="font-size:108%"> Sep.22th</b></h1> |
| − | <div style="padding-left:32px;">1.Gel electrophoresis showed that amplification of fragments was successfull.<br/> | + | <div style="padding-left:32px;"><li>Gel electrophoresis showed that amplification of fragments was successfull.</li> |
| − | 2.Ligated <i>13</i> and <i>19-15</i> via overlap PCR.<br/>
| + | <li>Ligated <i>13</i> and <i>19-15</i> via overlap PCR.</li> |
| − | 3.This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.<br/>
| + | <li>This PCR worked well and the products were extracted from the gel using a Agarose Gel DNA Extration Kit.</li> |
| − | 4.Restriction digest on pCPC-3031-Ni with Sac I.<br/>
| + | <li>Restriction digest on pCPC-3031-Ni with Sac I.</li> |
| | | | |
| | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"><br/> | | <img align="center" src="https://static.igem.org/mediawiki/2016/8/89/Igem-6803-week6-2.jpg" alt="Igem-6803-week6-1" width="300px"><br/> |
| | | | |
| − | 5.The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.<br/>
| + | <li>The fragment <i>13-19-15</i> was ligated onto T vector and transformed into E.coli via heat shock.</li> |
| | | | |
| | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" ><br/> | | <img align="center" src="https://static.igem.org/mediawiki/2016/0/06/Igem-6803-week6.jpg" alt="Igem-6803-week6-2" width="300px" ><br/> |
| Line 450: |
Line 450: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.23th</b></h1> | | <b><h1 style="font-size:108%"> Sep.23th</b></h1> |
| − | <div style="padding-left:32px;">1.A colony PCR of pT-13-19-15 was performed with 12 colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR of pT-13-19-15 was performed with 12 colonies.</li> |
| − | 2.Two of these colonies containing pT-13-19-15 were used to inoculate overnight cultures.<br/>
| + | <li>Two of these colonies containing pT-13-19-15 were used to inoculate overnight cultures.</li> |
| − | 3.<i>13-19-15</i> gene fragment was phosphorylated.<br/>
| + | <li><i>13-19-15</i> gene fragment was phosphorylated.</li> |
| − | 4.Phosphorylated <i>13-19-15</i> was ligated into pCPC-3031-Ni and transformed into E.coli via heat shock.<br/>
| + | <li>Phosphorylated <i>13-19-15</i> was ligated into pCPC-3031-Ni and transformed into E.coli via heat shock.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 459: |
Line 459: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.24th</b></h1> | | <b><h1 style="font-size:108%"> Sep.24th</b></h1> |
| − | <div style="padding-left:32px;">Plasmids pT-13-19-15 were isolated using a miniprep kit.<br/> | + | <div style="padding-left:32px;"><li>Plasmids pT-13-19-15 were isolated using a miniprep kit.</li> |
| | </div> | | </div> |
| | <br/> | | <br/> |
| Line 493: |
Line 493: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.25th</b></h1> | | <b><h1 style="font-size:108%"> Sep.25th</b></h1> |
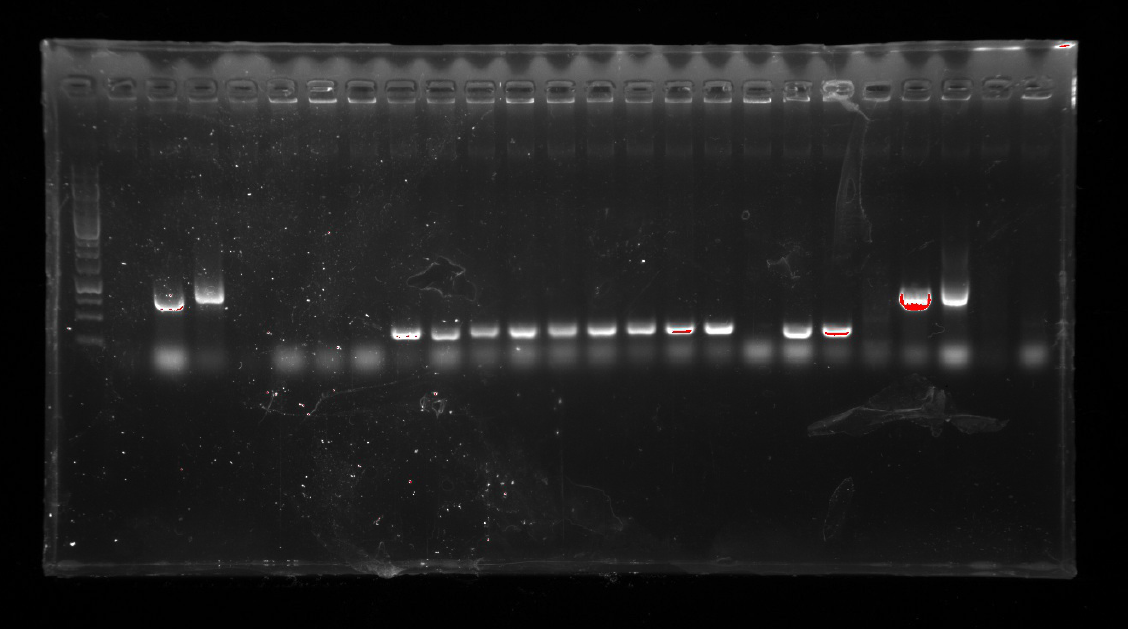
| − | <div style="padding-left:32px;">1.A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.<br/> | + | <div style="padding-left:32px;"><li>A colony PCR of pCPC-3031-Ni-13-19-15 was performed with 12 colonies.</li> |
| − | 2.Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.<br/>
| + | <li>Three colonies were proved to be true. And two of them were used to inoculate overnight cultures.</li> |
| | | | |
| | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" ><br/> | | <img align="center" src="https://static.igem.org/mediawiki/2016/2/2d/Igem-6803-week6-3.png" alt="Igem-6803-week6-3" width="800" height="533" ><br/> |
| Line 503: |
Line 503: |
| | | | |
| | <b><h1 style="font-size:108%"> Sep.26th</b></h1> | | <b><h1 style="font-size:108%"> Sep.26th</b></h1> |
| − | <div style="padding-left:32px;">Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.<br/> | + | <div style="padding-left:32px;"><li>Plasmids pT-13-19-15 were handed in for sequencing, which confirmed its correctness.</li> |
| | <b>The sequencing results for them were correct.</b> | | <b>The sequencing results for them were correct.</b> |
| | <br/> | | <br/> |