| Line 165: | Line 165: | ||
</div> | </div> | ||
| − | + | <br/> | |
<p style="font-size:18px">Data-processing.</p> | <p style="font-size:18px">Data-processing.</p> | ||
<div class="row"> | <div class="row"> | ||
Revision as of 13:17, 17 October 2016
Results
Overview
System Working Under Real World Conditions.
Synthetic biology aims at using biological part to construct different kinds of devices to realize different functions in real world. Experiments in the laboratory is idealized so it cannot represent the real world conditions all the time. In order to simulate the real world condition without taking the biological materials outside the lab, we can use PET products bought from the regular shop instead of the chemical shop. Considering the organisms are living with each other in real world, we applied the microbial consortium to simulate the real world condition as much as possible.
Using the Real World Substrate
Our project this year is about biodegradation of the widely used palstic PET. As a polymer, PET is hard to degrade and the degrading process is time-consuming. Due to the mild condition of the biodegradation reaction and the restriction of growing of organisms, it is even slower although it is more environmental friendly. Therefore, we use the substitute pNPA (p-nitrophenyl acetate), a kind of small molecular ester as the reagent of enzymatic reaction. However, pNPA is not what we really want to degrade, so we need to measure the PET degrading effects of our enzyme. Actually, pNPA is mainly used to screen for the mutated PETase gene because of the high requirement of efficiency of this step. When we obtain the mutants we want, we used them to degrade PET. In order to simulate the real world condition as much as possible, we used the PET film bought from the online plastic products shop instead of the test reagent bought from the chemical company. The film is originally used as packing material. We soak the cut film into the 75% ethanol solution and wash them with double distilled water before degrading experiments.


Using the Real World Simulated Microbial Consortium
The inspiration of the idea of microbial consortium comes from nature. Actually, bacteria never exist alone in our nature, they co-work and cooperate together to achieve an aim or live better in a special condition. Thinking from this point, we established a special bacteria consortium for this enzyme catalysis reaction in order to not only improve the degradation effect, but also simulate the real world condition. In this system ,we applied three kinds of organisms, Pseudomonas putida KT2440, Rhodococcus jostii RHA1 and Bacillus stubtilis 168 (or Bacillus stubtilis DB 104), to degrade the PET and the harmful product EG and TPA to carbon dioxide or generate biodegradable plastic PHA. We also decided to introduce the Cyanobacterial into the system in the future in order to make this system use only solar energy to work.

Experiment Data of Respective Section
Protein Engineering
Overview
We utilized the cell-free system to express the enzymes which had been modified in 22 different sites. Then we used the proteins successfully expressed to degrade PET. Our expected goal was to screen mutations with higher enzyme activities than the wild type PETase. .
Detailed results
After static reaction at 39℃ for 5 days, we detected the characteristic adsorption peak of the product ,MHET, which has no other characteristic adsorption peak except in 260nm.

Fig.1. Spectral scan for the degradation product MHET

Fig.2.Spectral scan for the degradation product MHET(Samples with no other absorption peak except in 260nm)
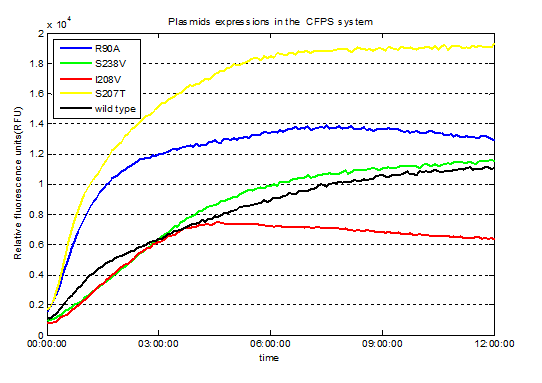
Fig.3. Screened plasmids expressions in the CFPS system

Fig.4. Relative activities of enzymes.
Data-processing.
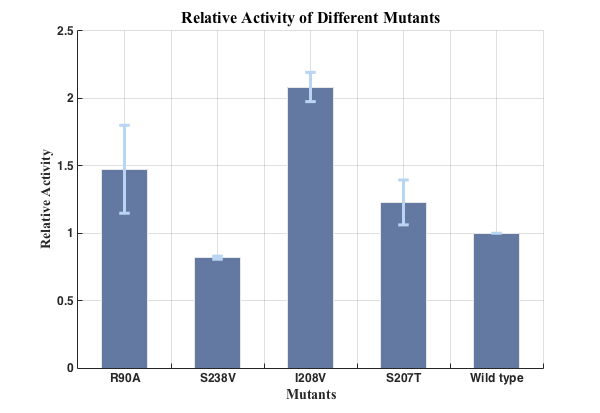
Fig.5. Relative activities of enzymes(n=3).
Summary
We successfully screened two mutants (I208V & R90A ) with higher enzyme activity by site-directed mutation.
You can find our parts page here:I208V
R90A
Microbial Consortium
xxx
R-R system
1.Results of inclusion body based reporting system.
The red fluorescence can be observed by bare eyes. We set up four groups:
A. No E.coli.
B. E.coli with empty plasmid pUC19.
C. E.coli with pUC19+CpxR-RFP.
D. E.coli with pUC19+CpxR-RFP+PETase gene.
The result is as the following picture. (After centrifugation with the speed of 12000rpm for 1min)

Fig.1. Directed observed fluorescence of inclusion body based reporting system. (From left to right: A, B, C, D)
We found that the group with part BBa_K339007 and PETase gene showed deepest red in all of the four groups. The group with only part BBa_K339007 also showed a little red, we speculate that this might be caused by the basic expression of some genes in E.coli and some of the expression products formed inclusion bodies. It might also be caused by the basic expression of the RFP gene because the CpxR promoter might also, though not so strongly, start the transcription even without the induction of CpxR protein. The group with empty plasimd did not showed any red color and the group with no E.coli cultured did not have any sediment at the bottom of tube.
2.Results of cell lysis effect of ddpX gene.
We first did a experiment to measure the cell lysis effect of ddpX gene. We simply use the IPTG inducible T7 promoter to regulate the expression of ddpX gene. We totally set four groups:
1. E.coli wildtype.
2. E.coli wildtype added IPTG.
3. E.coli with ddpX gene, no induction.
4. E.coli with ddpX gene, IPTG added as induction.
Then we continuously measure the OD600 of the culture medium by 96-well Microplate Reader and draw the OD600-culturing time curve by Matlab. The graph is showed below.

Fig.2. OD600-culturing time curve of different groups of inclusion body induced cell lysis system.
From this image, we can see that in the first 5 hours, the OD600 of each group is almost the same because of the rich nutrition and the ddpX cannot take effect immidiately. However, after 5 hours, the group with ddpX transformed and IPTG induced showed the fastest decrease among all the groups. This is because of the cell lysis effect of ddpX. Comparing the group with ddpX gene and IPTG inducted with the group with wildtype bacterial and the same amount of IPTG added we can draw the conclusion that the ddpX gene can cause cell lysis significantly. However, the data of another two groups are a little strange. We think it is because of the basic expression of ddpX can partly hydrolyze the peptidoglycan in the cell wall and provide cells with D-Ala as carbon source. This is exactly the physical use the the ddpX gene when the cell is under starvation condition, see more details about this explanation in our experiment page. Therefore the group with ddpX gene but no induction can live longer and better when the nutritions are deficient.
2.Results of inclusion body based cell lysis regulation system.
Summary
1.We successfully applied and improved the previous part BBa_K339007 designed and constructed by the Group iGEM10_Calgary to construct our inclusion body based reporting system and inclusion body based cell lysis regulation system.
2.The red fluorescence can even be directly observed by bare eyes when we express the PETase gene in the E.coli which was transformed into the CpxR-RFP fragment.
3.When we replace the RFP gene with the ddpX gene, when the PETase gene was expressed, we detected the decrease of OD of culture medium, which showed the lysis of E.coli.
4.The TPA positive feedback system can be induced by TPA. We used RFP as reporting gene and found that the higher the TPA concentration is, the stronger the red fluorescence could be detected. We believe that if the RFP gene is replaced by our PETase gene, the expression of PETase gene can also be induced by the PET degradation product TPA.


