| Line 55: | Line 55: | ||
<h2><b>Protein Modification</b></h2> | <h2><b>Protein Modification</b></h2> | ||
| − | <p style="font-size:18px"> | + | <p style="font-size:18px"><h3>Plasmid construction and expression in CFPS system</h3> |
| + | Ligaion of digested pRset_CFP-1, digested CFP gene and digested PETase(I208V) gene in accordance with the 1:5:5 molecular ratio. The newly constructed plasmid is called pRset_CFP-1_PETase-I208V. | ||
| + | Than , the plasmids pRset_CFP-1_PETase-I208V was put into the CFPS(Cell-Free Protein Synthesis) to synthesis the enzymes we expected. | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | <div class="col-md-8"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/e/e6/T--Tianjin--part-I208Vpc.png" alt="desktop"> | ||
| + | <p style="font-size:15px"> | ||
| + | <br/> | ||
| + | Fig.1. Plasmid construction and expression in CFPS ststem. | ||
| + | </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <h3>Degradation data</h3> | ||
| + | After the enzymes were expressed in the system successfully, we used the proteins we got to degrade PET and detected the degradation product, MHET, which has no other characteristic adsorption peak except at 260nm. | ||
| + | |||
| + | <li>We detect the absorption peak of culture medium after culturing with PET film added for 5d. The absorption degree-wavelength curve is as follows.</li> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | <div class="col-md-8"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/thumb/7/7a/T--Tianjin--partdataI208V.png/800px-T--Tianjin--partdataI208V.png" alt="desktop"> | ||
| + | <p style="font-size:15px"> | ||
| + | <br/> | ||
| + | Fig.2.Multispectral scanning for the degradation product. | ||
| + | </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <li>We used the relative fluorescence units to represent the expression level of enzymes. </li> | ||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | <div class="col-md-8"> | ||
| + | <img src=" https://static.igem.org/mediawiki/2016/1/1d/T--Tianjin--part-I208Vex.png" alt="desktop"> | ||
| + | <p style="font-size:15px"> | ||
| + | <br/> | ||
| + | Fig.3.Expression of Plasmid pRset_CFP-1_PETase-I208V. | ||
| + | </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <li>Finally we got the relative activities of enzymes. </li> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | <div class="col-md-8"> | ||
| + | <img src=" https://static.igem.org/mediawiki/2016/2/2f/T--Tianjin--part-I208Vbar.png" alt="desktop"> | ||
| + | <p style="font-size:15px"> | ||
| + | <br/> | ||
| + | Fig.3.Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V. | ||
| + | </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<h2><b>Microbial Consortia</b></h2> | <h2><b>Microbial Consortia</b></h2> | ||
<p style="font-size:18px">xxxxxxxxxx</p> | <p style="font-size:18px">xxxxxxxxxx</p> | ||
Revision as of 10:06, 18 October 2016
Proof
Introduction
Our project mainly consists of three integrated sections--Protein Modification, Microbial Consortium and Reporting-Regulation System. In each section there are many concepts related to our experiment design. Our project focused on proving these concepts by constructing new parts. Our proof of concepts are divided into three sections respectively belong to the three sections of our whole project. The detailed proof method and relative constructed devices are as follows.
Protein Modification
Plasmid construction and expression in CFPS system
Ligaion of digested pRset_CFP-1, digested CFP gene and digested PETase(I208V) gene in accordance with the 1:5:5 molecular ratio. The newly constructed plasmid is called pRset_CFP-1_PETase-I208V. Than , the plasmids pRset_CFP-1_PETase-I208V was put into the CFPS(Cell-Free Protein Synthesis) to synthesis the enzymes we expected.
Fig.1. Plasmid construction and expression in CFPS ststem.
Degradation data
After the enzymes were expressed in the system successfully, we used the proteins we got to degrade PET and detected the degradation product, MHET, which has no other characteristic adsorption peak except at 260nm.
Fig.2.Multispectral scanning for the degradation product.
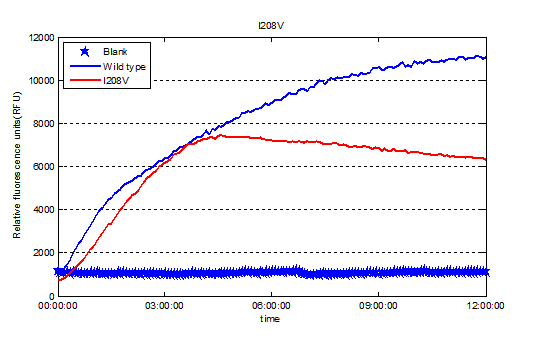
Fig.3.Expression of Plasmid pRset_CFP-1_PETase-I208V.

Fig.3.Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V.
Microbial Consortia
xxxxxxxxxx
R-R System
Introduction
R-R (Reporting and Regulation)system is constructed in order to report the expression of PETase gene and regulate the expression process. In this section there are two main concepts we need to prove. The first is whether the ddpX gene can cause cell lysis and the other is whether the ddpX gene can cause cell lysis under the CpxR promoter when inclusion body form led by the overexpression of PETase gene. We constructed the part BBa_K2110008 to prove the validity of this concept.
Constructing Process
This part is modified and improved from the former part BBa_K339007 by changing the original mRFP gene to the novel ddpX gene (Part: BBa_K2110004). The ddpX gene was obtained by colony PCR of E.coli and was verified by sequencing. Considering there is no restriction endonuclease cutting site among the subparts, we used PCR to amplify the CpxR promoter-RBS sequence and linked it with the plasmid backbone and the ddpX gene. Then the new part was obtained.
Proof Result
After we constructed the new biobrick, we firstly used PCR to amplify the whole biobrick and linked it to the recombinant plasmid pUC19 already with PETase gene in it. Then we transfromed the plasmid into E.coli and induced the expression of PETase gene. Then we measured the OD600 of the culture medium. The result is showed below.



