Results of CFPS
Overview
We utilized the cell-free system to express the enzymes which had been modified in 22 different sites. Then we used the proteins successfully expressed to degrade PET. Our expected goal was to screen mutations with higher enzyme activities than the wild type PETase. .
Detailed results

Fig.1. Spectral scan for the degradation product MHET

Fig.2. Spectral scan for the degradation product MHET (no other peak except in 260nm)
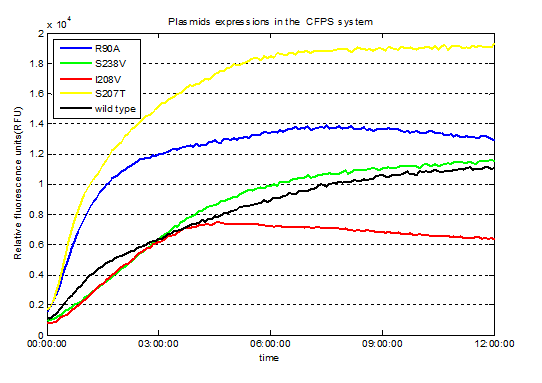
Fig.3. Screened plasmids expressions in the CFPS system

Fig.4. Relative activities of enzymes
Summary
Basically, we utilized the cell-free system to express the enzymes which had been modified in 22 different sites. Besides, we added a fluorescet protein, CFP, before the enzyme. And there is a flexible linker, GGGGSGGGGS , between them. So that we could detect the expression of enzymes by detecting expression of the fluorescent protein with a fluorescence readout instrument, for example, a microplate reader. We conceived that with this method we could acquire the best modifications by screening them in a high-throughput way. Then we used the proteins we got to degrade PET.

Fig.3.The expression vector in CFPS system
How to characterize the degradation velocity is the main problem in our scheme. We analyzed the experiment consequences in two ways. For the first one, we rendered the enzymes degrade pNPa, a general substituent for the detection of PET. Then we measured the absorbance of pNP in the optical density of 400 nanometers, which is the degrading product of pNPa. For the second one, we detected the absorbance of MHET in the optical density of 260 nanometers, which is the product in the first step of PET degradation.

Fig.4.Steps for integrated expression and activity screening of enzymes


