Advantages and Limitations
Throughout the development of BabblED we have tried to target a number of key areas in DNA storage to improve upon which are outlined below.
Speed of DNA Synthesis
One of the benefits of our system is the rapidity of DNA assembly. Because we aimed to develop a system that avoids de novo DNA synthesis, we also avoid all the drawbacks that come with it. This graph compares the speed of DNA assembly in DNA bases assembled per minute for BabbleBrick/BabbleBlock assembly using the protocol we designed, versus the ADTBio solid-phase oligonucleotide synthesis protocol. (Reference: http://www.atdbio.com/content/17/Solid-phase-oligonucleotide-synthesis#Detritylation-of-the-support-bound-3-nucleoside)

Cost comparison
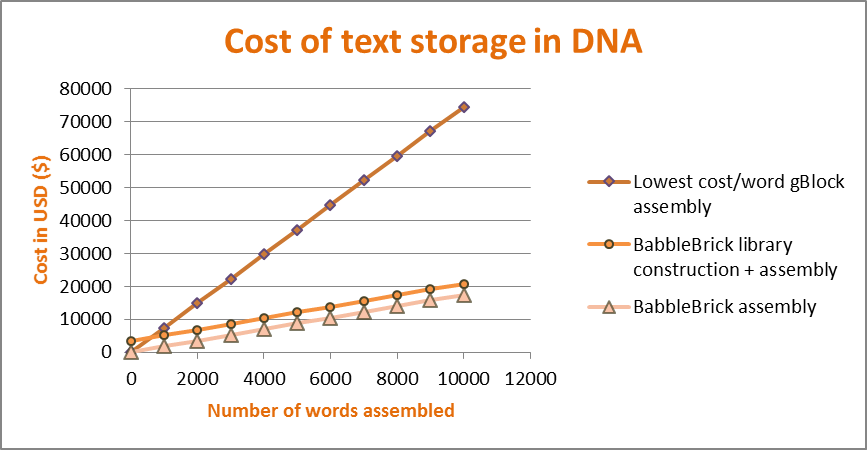
This graph shows the cost comparison for DNA synthesis using gBlocks (cheapest per base pair synthesised) versus our modular system for BabbleBrick assembly. We have assumed that we constructed a library of 1000 words. Even though library construction adds a small, but steep, increase in the cost, subsequent assembly is much cheaper, and we make our money back after assembling 500 words. Subsequent assemblies will be much cheaper, as our library is reusable and therefore we don’t need to construct a new one.
We have assumed that we construct a library of 1000 unique words, and used NEB polyT magnetic beads and the NEB quick ligase kit for assembly. We compared it against ordering a ready-made de novo synthesised gBlock sequence encoding the same piece of text.
Every Scientist things their project is amazing and world-changing. Us included! However, we appreciate that all projects have limitations due to their design, implementation, lab equipment, finance, time..
Limitations of our Project
As an iGEM project, our main limitation was time. Ideally we would have like to done a lot more preliminary lab work to test optimal anchor binding, optimal volumes of liquids for assembly, a different method of assembly (one-pot), to name a few. We would also like to have access to a minion to test its ability to sequence our data, as that is what we would propose to use if our information was stored in a non-lab commercial setting.
Limitations of our Design

Our design is modular, novel, and importantly cheaper than de novo synthesis, which was our main goal. However there are constant improvements we can be making to it. One of the main limitations of the design is the waste in using excess reagents. One way to combat this would be to use an automated system. An example set-up is shown below.
Another limitation is the number of BabbleBricks we can use per BabbleBlock. If the desired construct is to be re-inserted into a phytobrick then there is a limit to the number of BabbleBricks that can be assembled together. We have tried to target this limitation be incorporating an address. At the end of every BabbleBlock there is an address. This tags the construct with a code that can be identified by the Babbled decoding software. The address information places the construct in context of other constructs. Eg which sentence in a paragraph it is, or which line of pixels on a picture. This makes assembly of encoded data from many constructs possible.




