Design
General Design
To develop a novel cell-free platform for the low-cost, high-efficient and visualized detection of serum miRNAs, two essential systems, namely RCA based DNA amplification system, and dCas9 conjugated split-reporting system, were combined, modified, and then assessed in our project.
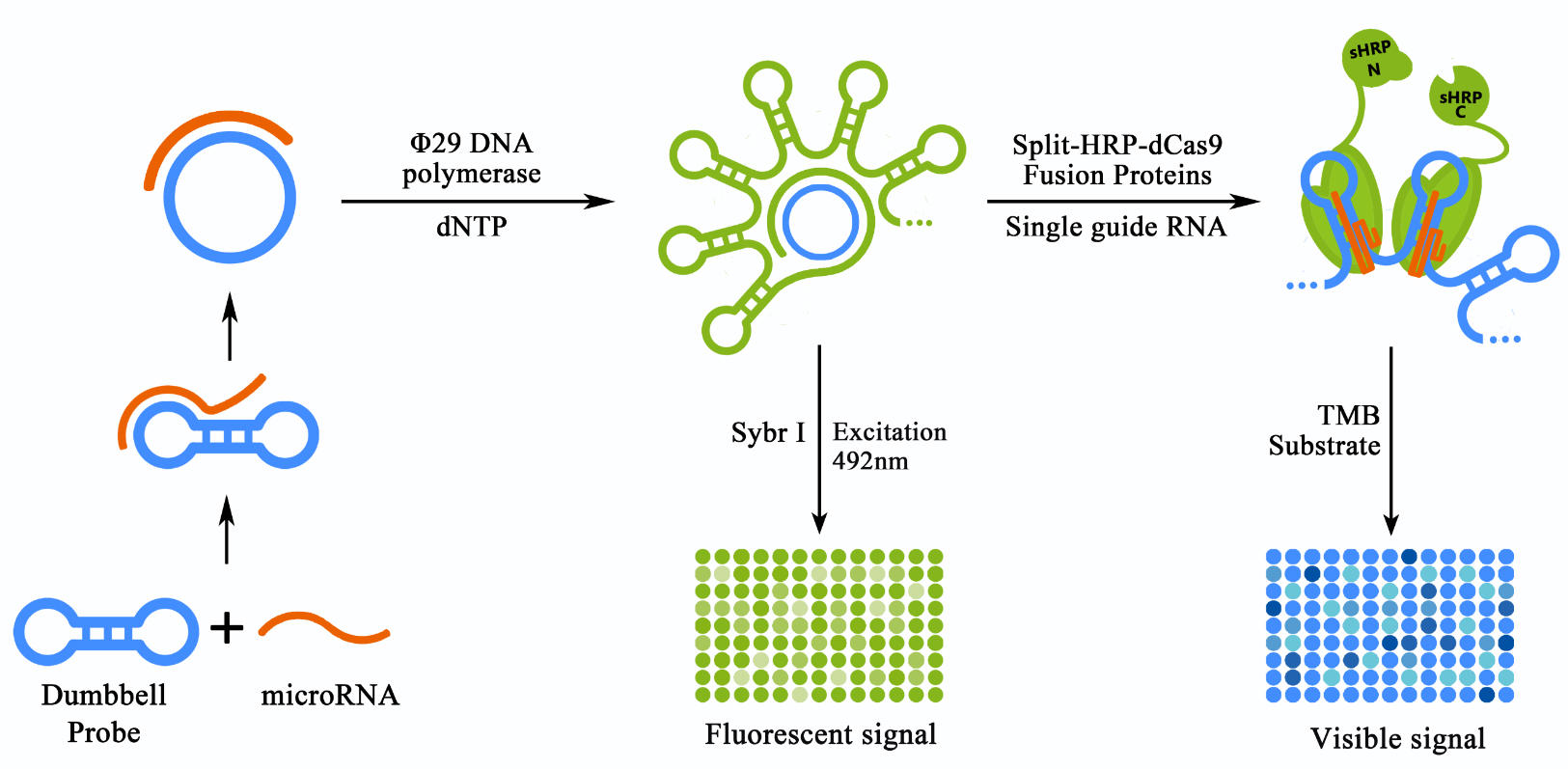
The first system, RCA based DNA amplification system, was introduced to primarily amplify the input miRNA signal with a high specificity under an isothermal and moderate condition. Specifically, a dumbbell shaped probe containing a tunable toehold domain on its loop was custom designed and prepared for the detection of a specific target miRNA. Target miRNAs could bind with the toehold domain, and then trigger the toehold-mediated strand displacement (TMSD) process resulting in a switch of the probe from the dumbbell shaped form into a circular form (termed as initiated probe, or iprobe in brief). The iprobe could then be used as the template for the subsequent RCA reaction (Figure 1, left panel). A mismatched miRNA, however, would fail to trigger the TMSD due to the resistance of the stabilized dumbbell structure, thus producing no amplification products. Once RCA products were produced, a traditional Sybr I based fluorescence assay could be conducted to assess the effectiveness of RCA based DNA amplification system. A fluoresce microplate reader would be needed for such matters (Figure 1, middle panel).
Figure 1. Schematic representation of the detection process of our scheme.
The miRNA in the sample can bind with the pre-designed dumbbell probe. Once bond, the chain-replace reaction would be activated and the dumbbell shaped probe would be transformed into a circular structure, thus allowed the initiation of the rolling cycle DNA amplification (RCA) process. The product of RCA reaction can be detected directly through Sybr I staining and a fluorescent plate reader. The RCA product were to be bound by sgRNA guided dCas9 proteins fused with split-HRP reporters, thus produced a visible and stronger signal by producing TMB diamine from TMB substrate.
In order to achieve the further amplification and visualization of the RCA output signal, the dCas9 conjugated split-reporting system was then introduced into our scheme. The fusion proteins of dCas9 and split-HRP fragments, namely sHRP-N-dCas9 (N-sHdC) and sHRP-C-dCas9 (C-sHdC) could be obtained from genetically modified E. coli strains containing relevant expression plasmids. With the guidance of sgRNA, N-sHdC proteins and C-sHdC proteins would be able to bind randomly to the numerous double-strand loci on the RCA product (Figure 2, right panel). For all those bound to loci that were close enough, split-HRP fragments would interact with each other and HRP enzyme activity would be retained. Building on this, HRP enzyme activity could then be determined by adding substrates such as 3,3',5,5'-Tetramethylbenzidine (TMB) with a visual output signal.
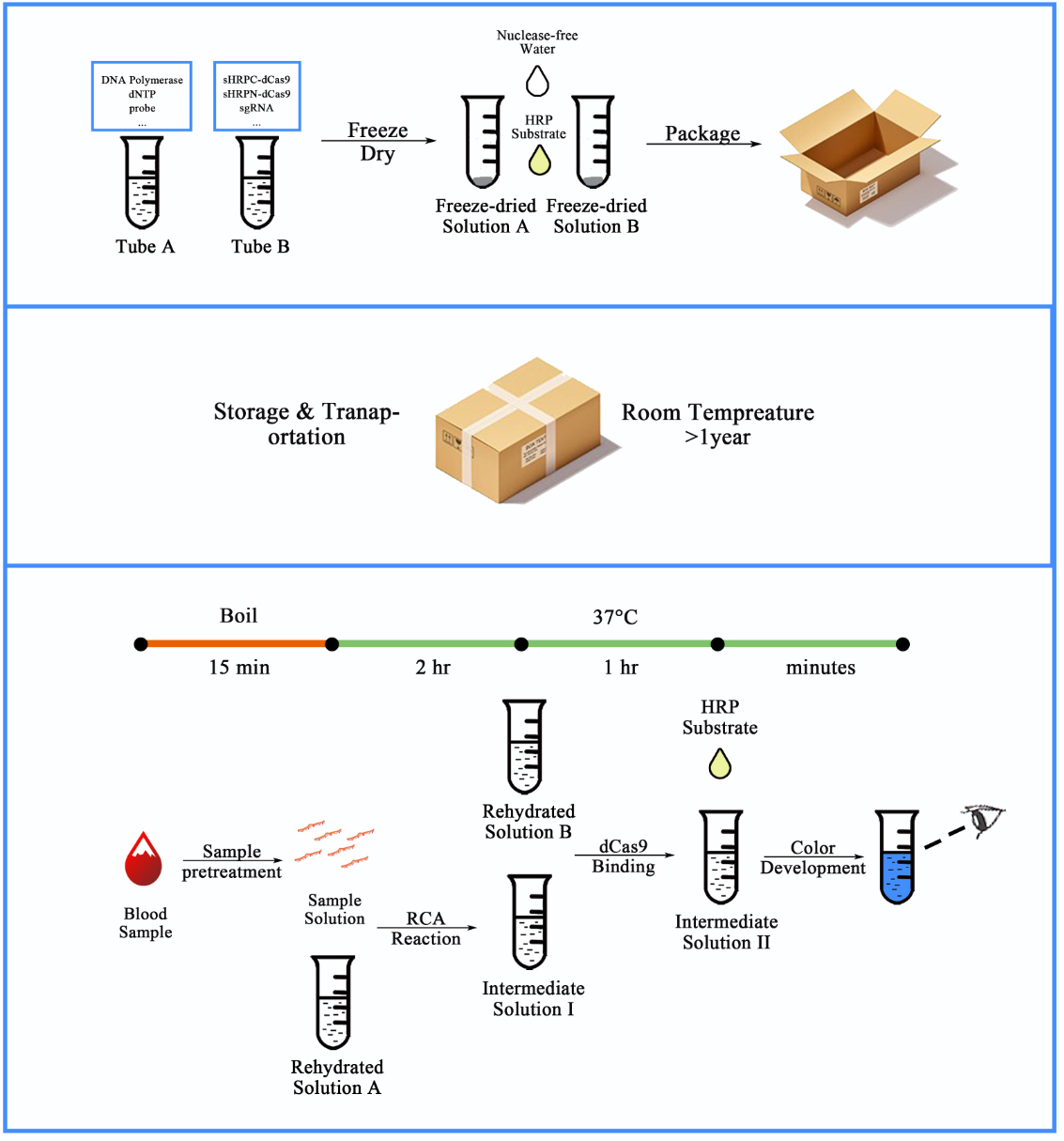
Figure 2. Workflow for our CRISPR-based blood-microRNA detection system.
The synthesized and sealed dumbbell probe together with other RCA related materials can be embedded into tube A, and purified fusion proteins of dCas9 and split-HRP fragments together with other dCas9-binding related materials can be embedded into tube B. The reagents in both tubes can be freeze-dried to maintain stable in room temperature for a relatively long time (>1 year). When entering the detection process, serum samples were pre-treated by boiling for 15 min to expose the miRNAs from molecule complexes and exosomes. After a series of simple operations, the amount of the specific RNA could be indicated by the color depth from the colorless to the dark blue.
Since most proteins could remain stable under normal storage conditions if freeze dried, and retain their activity after rehydrated. Our system could then be developed into a kit that contains freeze-dried components together with other liquid components that is stable in regular storage conditions, such as TMB substrate and DEPC treated water (Figure 2, upper and middle panel). Meanwhile, with its advantage in visualized outputs and moderate temperature (37℃) detection process, such kit would be able to be deployed in low-resource settings and dramatically lower the cost of and technical barrier for wider cancer scanning and early detection.
Prototype: serum let-7a detection system
To validate and demonstrate our design, miR let-7a, as an important serum biomarker for non-small cell lung cancer, was chosen as the target miRNA. Previously, miR let-7a has been reported to be down-regulated for 20%-40% in serum samples from NSCLC patients compared to healthy people 1.
To produce miR let-7a for further detection, we put the sequence of let-7a under control of a T7 promoter. The plasmid was linearized at the point right after let-7a sequence and in vitro transcription kit was then used for the transcription of let-7a.
AS A PROOF OF CONCEPT, let-7a was diluted in DEPC-treated water on various concentrations to assess the reliability, sensibility and specificity of our scheme. To begin with, four different probes were designed to be probe candidates for the RCA reaction based on let7a sequence and probe design principles. Once they have been synthesized and purified, RCA reactions with 10nM let-7a input were performed against all four probes to select the optimal probe for further test. By using this probe, the sensibility and specificity of RCA reaction were then determined with sybr I fluorescence assay. Moreover, N-sHdC and C-sHdC protein were expressed and purified from E.coli, and subsequently used for the dCas9 binding process together with an in vitro expressed sgRNA. TMB substrate was used to test the HRP activity. (See proof of concept page for more details)
FOR FURTHER DEMONSTRATION OF OUR PROJECT, let-7a was dissolved in 7% mixed human serum collected from 50 healthy volunteers. Similarly, sybr I fluorescence assay and HRP activity assay was conducted to verify the reliability, sensibility and specificity of our scheme in stimulated clinical samples. MOREOVER, serum samples collected from NSCLC patients and healthy volunteers were also tested after different pretreatment for further demonstration of our scheme. (See demonstration page for more details)
Reference
1 Jeong, H. C. et al. Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Mol Med Rep 4, 383-387, doi:10.3892/mmr.2011.430 (2011).