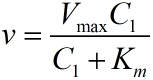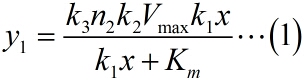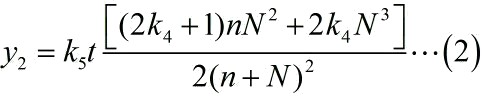Zhuchushu13 (Talk | contribs) |
Zhuchushu13 (Talk | contribs) |
||
| Line 88: | Line 88: | ||
action=raw&ctype=text/css" /> | action=raw&ctype=text/css" /> | ||
| − | <link rel="stylesheet" href="/Team:NUDT_CHINA/CSS? | + | <link rel="stylesheet" href="/Team:NUDT_CHINA/CSS? |
| − | /></head> | + | action=raw&ctype=text/css" /></head> |
<!-- body --> | <!-- body --> | ||
| Line 119: | Line 119: | ||
href="#">PROJECT</a></li> | href="#">PROJECT</a></li> | ||
<li class="nav-global__item trigger "> | <li class="nav-global__item trigger "> | ||
| − | <a class="nav-global__link" title="" href="#">PARTS</a></li> | + | <a class="nav-global__link" title="" |
| + | |||
| + | href="#">PARTS</a></li> | ||
<li class="nav-global__item trigger "> | <li class="nav-global__item trigger "> | ||
<a class="nav-global__link" title="" | <a class="nav-global__link" title="" | ||
| Line 208: | Line 210: | ||
<div class="tai__image-group-wrap large "> | <div class="tai__image-group-wrap large "> | ||
<div class="tai__image-group single-image"> | <div class="tai__image-group single-image"> | ||
| − | <!--a href="https#/support/donate/annual | + | <!--a href="https#/support/donate/annual- |
| − | now/" class="lazyload-child"> | + | fund/donate-now/" class="lazyload-child"> |
<img src="" /></a--> | <img src="" /></a--> | ||
</div> | </div> | ||
| Line 248: | Line 250: | ||
<div class="tai__image-group-wrap large "> | <div class="tai__image-group-wrap large "> | ||
<div class="tai__image-group single-image"> | <div class="tai__image-group single-image"> | ||
| − | <!--a href="https#/support/donate/annual | + | <!--a href="https#/support/donate/annual- |
| − | now/" class="lazyload-child"> | + | fund/donate-now/" class="lazyload-child"> |
<img src="" /></a--> | <img src="" /></a--> | ||
</div> | </div> | ||
| Line 271: | Line 273: | ||
<div class="tai__image-group-wrap large "> | <div class="tai__image-group-wrap large "> | ||
<div class="tai__image-group single-image"> | <div class="tai__image-group single-image"> | ||
| − | <!--a href="https#/support/donate/annual | + | <!--a href="https#/support/donate/annual- |
| − | now/" class="lazyload-child"> | + | fund/donate-now/" class="lazyload-child"> |
<img src="" /></a--> | <img src="" /></a--> | ||
</div> | </div> | ||
| Line 294: | Line 296: | ||
<div class="tai__image-group-wrap large "> | <div class="tai__image-group-wrap large "> | ||
<div class="tai__image-group single-image"> | <div class="tai__image-group single-image"> | ||
| − | <!--a href="https#/support/donate/annual | + | <!--a href="https#/support/donate/annual- |
| − | now/" class="lazyload-child"> | + | fund/donate-now/" class="lazyload-child"> |
<img src="" /></a--> | <img src="" /></a--> | ||
</div> | </div> | ||
| Line 336: | Line 338: | ||
<div class="tai__image-group-wrap large "> | <div class="tai__image-group-wrap large "> | ||
<div class="tai__image-group single-image"> | <div class="tai__image-group single-image"> | ||
| − | <!--a href="https#/support/donate/annual | + | <!--a href="https#/support/donate/annual- |
| − | now/" class="lazyload-child"> | + | fund/donate-now/" class="lazyload-child"> |
<img src="" /></a--> | <img src="" /></a--> | ||
</div> | </div> | ||
| Line 364: | Line 366: | ||
<div class="tai__image-group-wrap large "> | <div class="tai__image-group-wrap large "> | ||
<div class="tai__image-group single-image"> | <div class="tai__image-group single-image"> | ||
| − | <!--a href="https#/support/donate/annual | + | <!--a href="https#/support/donate/annual- |
| − | now/" class="lazyload-child"> | + | fund/donate-now/" class="lazyload-child"> |
<img src="" /></a--> | <img src="" /></a--> | ||
</div> | </div> | ||
| Line 377: | Line 379: | ||
</div> | </div> | ||
</nav> | </nav> | ||
| − | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen | + | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen |
| − | (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } } | + | and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } } |
#feature{ | #feature{ | ||
| Line 437: | Line 439: | ||
bh__video" style="height:300px;"> | bh__video" style="height:300px;"> | ||
| − | <div class="bh__background bh__background-color "></div> | + | <div class="bh__background bh__background-color |
| + | |||
| + | "></div> | ||
<div class="bh__content-wrap" style="height:120px;"> | <div class="bh__content-wrap" style="height:120px;"> | ||
<div class="bh__content "> | <div class="bh__content "> | ||
| Line 510: | Line 514: | ||
| − | + | ||
| − | + | ||
<!--标题--> | <!--标题--> | ||
| Line 524: | Line 527: | ||
model is created to evaluate the effectiveness of initial design, and offers | model is created to evaluate the effectiveness of initial design, and offers | ||
| − | guidelines about how the system can (or must) be improved. (You can go PROJECT | + | guidelines about how the system can (or must) be improved. (You can go<a |
| + | |||
| + | href="/Team:NUDT_CHINA/Design"> PROJECT.page </a>to see more.) The core idea is | ||
| − | + | to simulate the process of producing the | |
signal which can be detected, and draw a conclusion by obtaining the | signal which can be detected, and draw a conclusion by obtaining the | ||
| Line 555: | Line 560: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | RCA | ||
model is based on the Michaelis-Menten equation. We can obtain the relationship | model is based on the Michaelis-Menten equation. We can obtain the relationship | ||
| Line 572: | Line 579: | ||
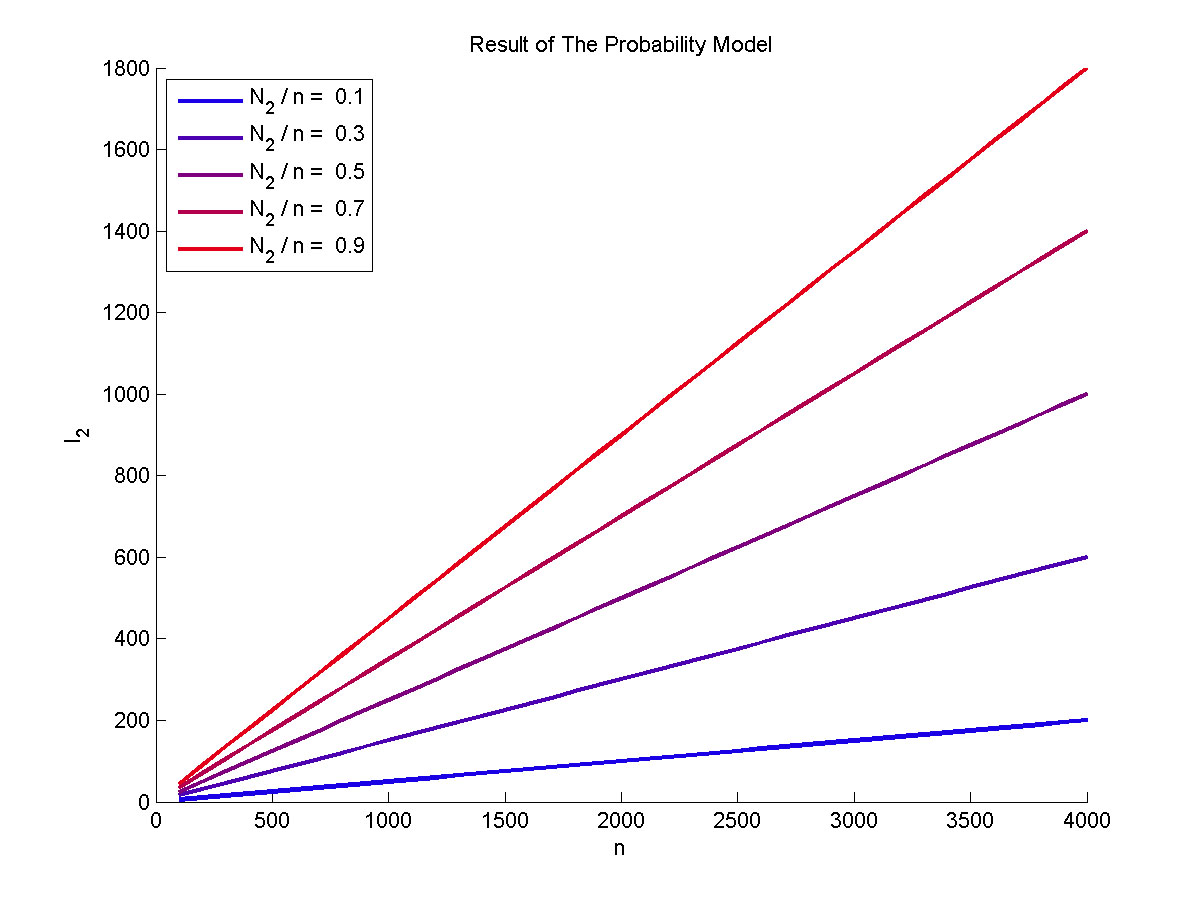
<span style="line-height:2;font-family:Perpetua;font-size:18px;">In the | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In the | ||
| − | signal detection model, we combine a probability model with the kinetic equation | + | signal detection model, we combine a probability model with the kinetic |
| + | |||
| + | equation | ||
of enzymatic reaction, so we can obtain the relationship between the number of | of enzymatic reaction, so we can obtain the relationship between the number of | ||
| Line 589: | Line 598: | ||
integrating the two models, we can theoretically predict the impacts of the | integrating the two models, we can theoretically predict the impacts of the | ||
| − | molecule number of proteins on the signal to noise ratio and explained the trend | + | molecule number of proteins on the signal to noise ratio and explained the |
| + | |||
| + | trend | ||
of the signal intensity with the change of the concentration of miRNA in our | of the signal intensity with the change of the concentration of miRNA in our | ||
| Line 651: | Line 662: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">We | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We | ||
| − | assume that the enzymatic activity remains unchanged with time under the premise | + | assume that the enzymatic activity remains unchanged with time under the |
| + | |||
| + | premise | ||
of excessive amount of enzymes or a short-time reaction.</span> | of excessive amount of enzymes or a short-time reaction.</span> | ||
| Line 769: | Line 782: | ||
windowtext 1.0pt;"> | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
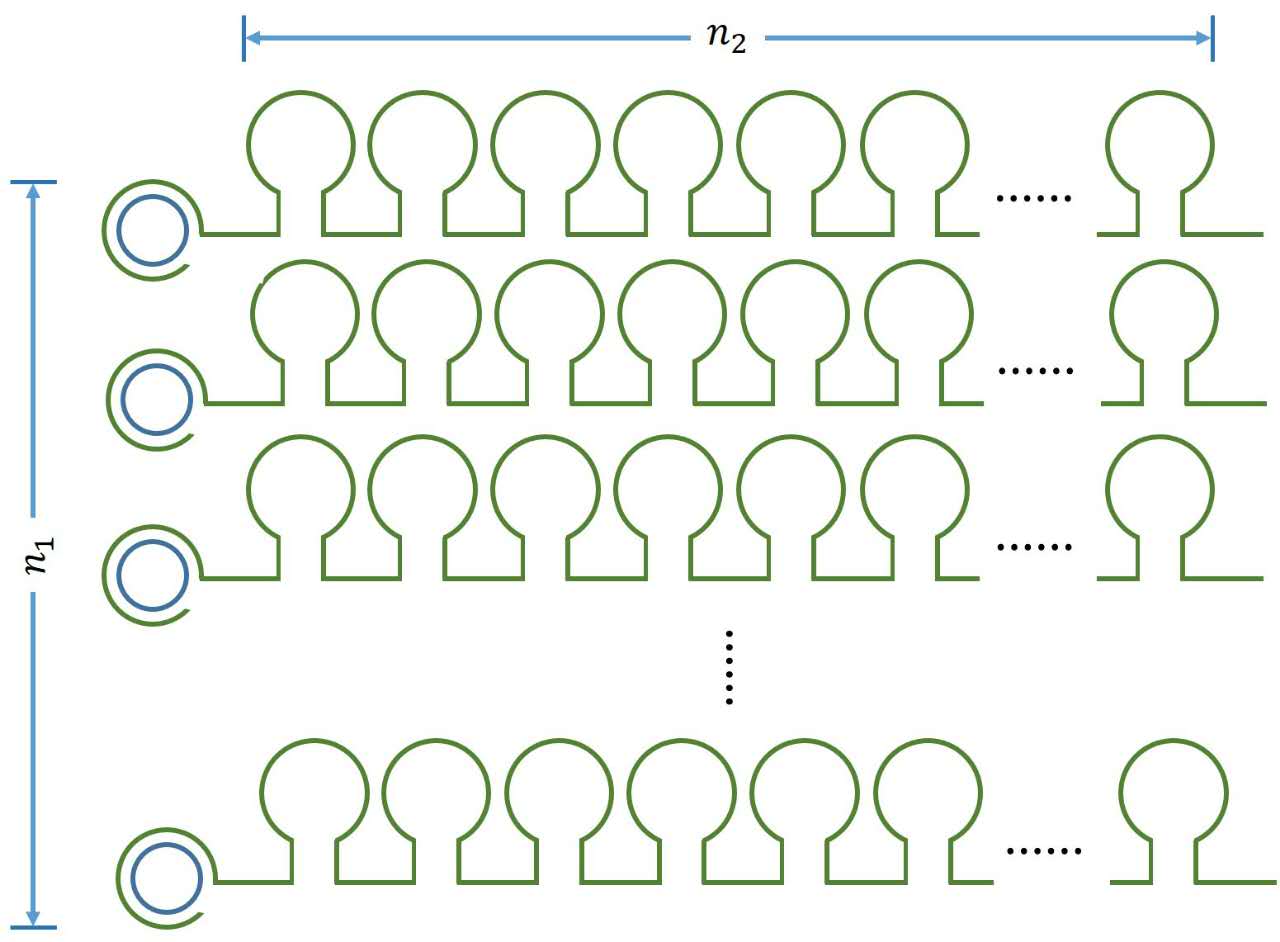
| − | <a name="OLE_LINK8"></a><span>A constant | + | <a name="OLE_LINK8"></a><span>A |
| + | |||
| + | constant | ||
representing | representing | ||
| Line 855: | Line 870: | ||
<a | <a | ||
| − | name="OLE_LINK9"></a><i><span>n<sub>1</sub></span></i><i><sub><span></span></ | + | name="OLE_LINK9"></a><i><span>n<sub>1</sub></span></i><i><sub><span></span></su |
| + | |||
| + | b | ||
></i> | ></i> | ||
| Line 1,198: | Line 1,215: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, | ||
| − | RCA is an enzymatic reaction, the reaction equation is written as: iprobe+ | + | RCA is an enzymatic reaction, the reaction equation is written as: iprobe+dNTP |
| + | |||
| + | → | ||
iprobe-dN+PPi, under the premise of excessive amount of enzymes and dNTPs, the | iprobe-dN+PPi, under the premise of excessive amount of enzymes and dNTPs, the | ||
| Line 1,235: | Line 1,254: | ||
extending of each RCA production is related to the reaction time, not the | extending of each RCA production is related to the reaction time, not the | ||
| − | concentration of the iprobe. It is a linear relationship when we assume that the | + | concentration of the iprobe. It is a linear relationship when we assume that |
| + | |||
| + | the | ||
enzymatic activity remains unchanged with time, thus, the number of stem-loop | enzymatic activity remains unchanged with time, thus, the number of stem-loop | ||
| Line 1,539: | Line 1,560: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">The | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| − | relationship between the number of Stem-loop structures and the signal intensity | + | relationship between the number of Stem-loop structures and the signal |
| + | |||
| + | intensity | ||
under the premise of adding a certain amount of the fusion proteins can be | under the premise of adding a certain amount of the fusion proteins can be | ||
| Line 1,558: | Line 1,581: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The calculation of the constants | + | <span><span style="color:#7f1015"> The calculation of the constants |
| + | |||
| + | |||
</span></span><hr /> | </span></span><hr /> | ||
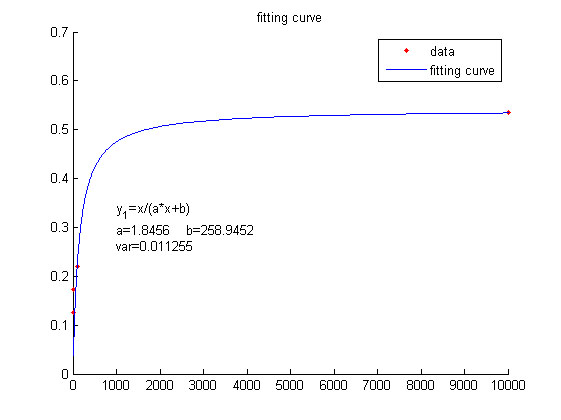
| Line 1,578: | Line 1,603: | ||
which refers to the number of stem-loop structures | which refers to the number of stem-loop structures | ||
in each RCA product, is stable under the premise of a certain reaction time. | in each RCA product, is stable under the premise of a certain reaction time. | ||
| − | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the equation | + | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the |
| + | |||
| + | equation | ||
can be | can be | ||
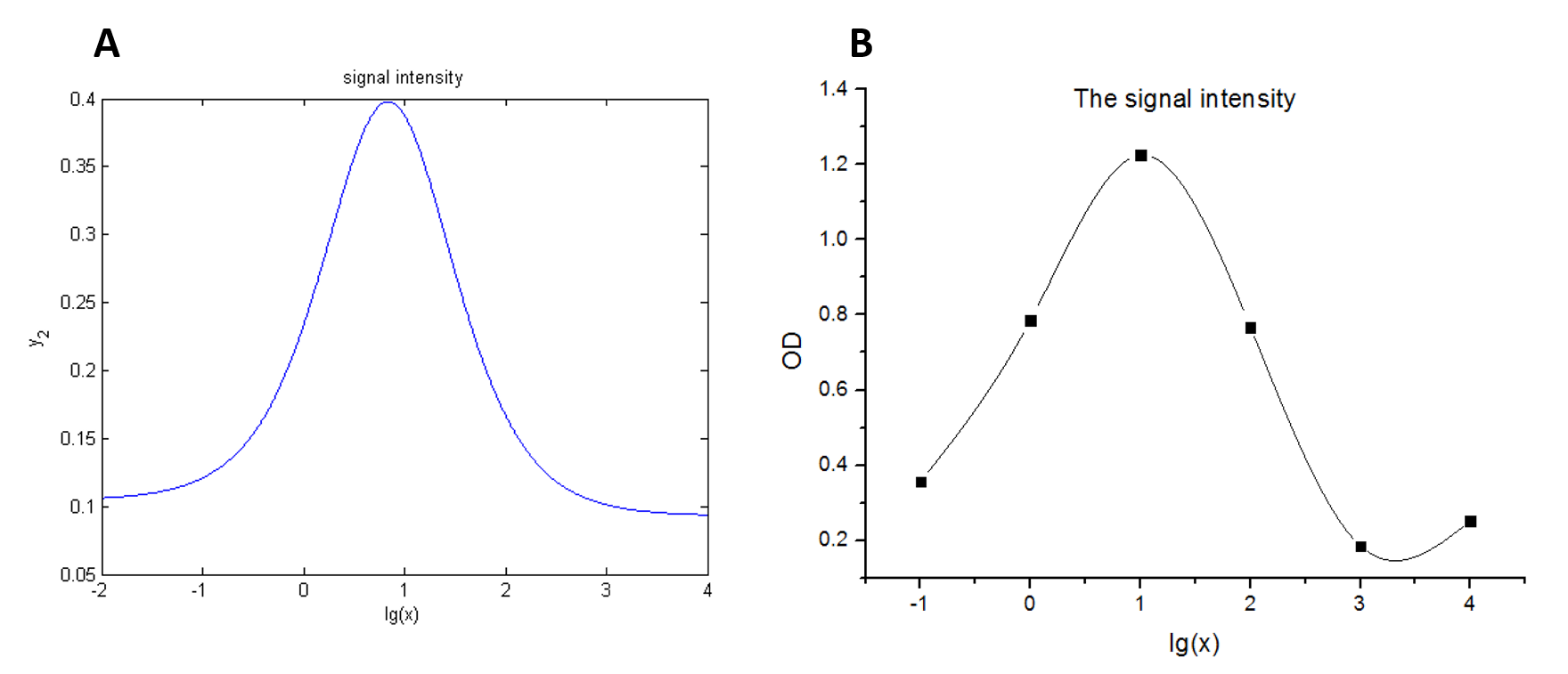
| Line 1,598: | Line 1,625: | ||
this equation to fit the data points obtained through experiments. (Figure 2. | this equation to fit the data points obtained through experiments. (Figure 2. | ||
| − | (D) in the RESULT. page) The fitting curve is shown below.</span> | + | (D) in the<a href="/Team:NUDT_CHINA/Result"> RESULT.page. </a>) The fitting |
| + | |||
| + | curve is shown below.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,679: | Line 1,708: | ||
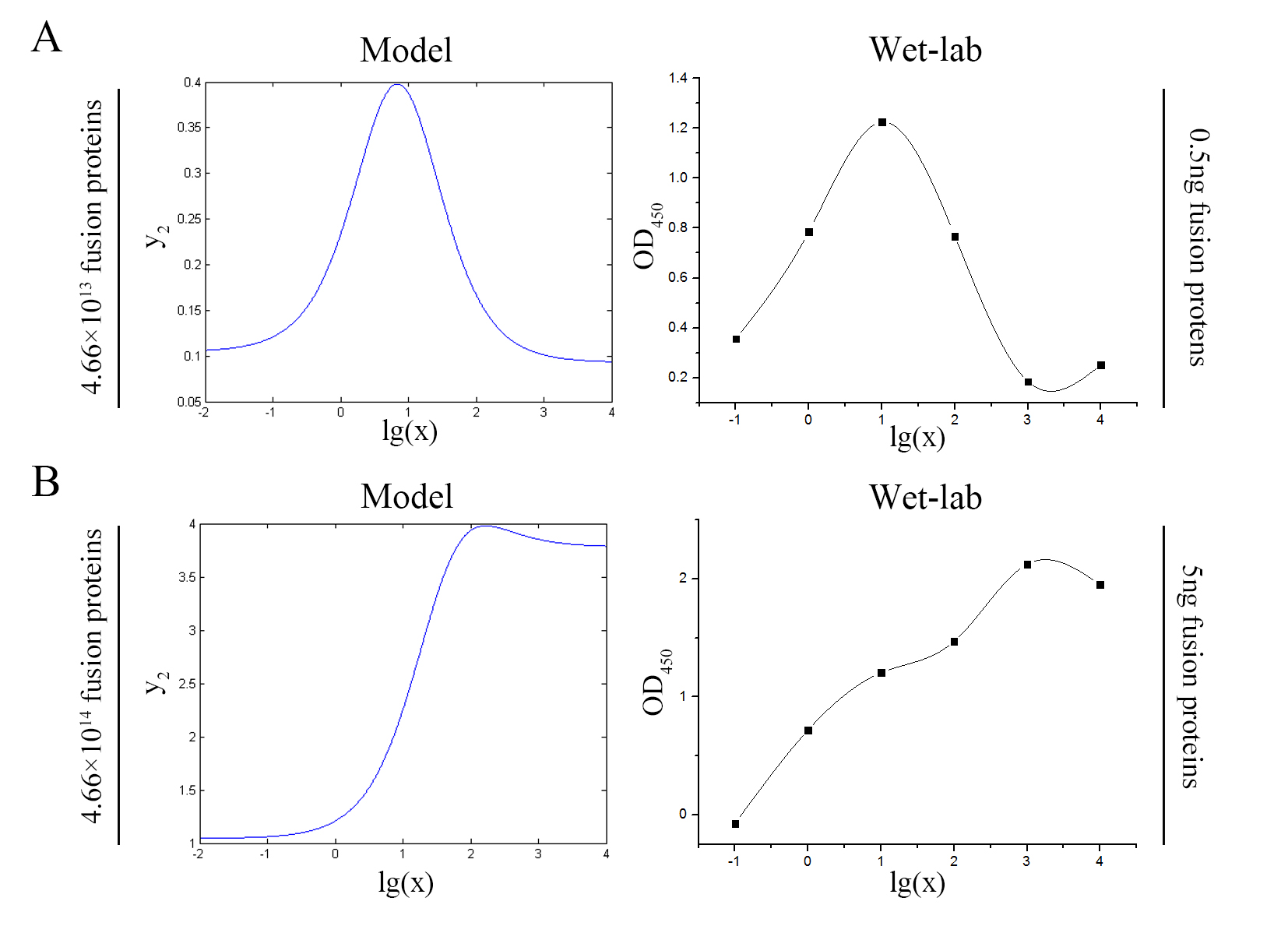
intensity (OD<sub>450</sub>) | intensity (OD<sub>450</sub>) | ||
| − | against miRNA concentration(pM) and additional amount of fusion proteins.</span> | + | against miRNA concentration(pM) and additional amount of fusion |
| + | |||
| + | proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,745: | Line 1,776: | ||
concentration of miRNA under the premise of adding a certain amount of the | concentration of miRNA under the premise of adding a certain amount of the | ||
| − | fusion proteins of dCas9 and split-HRP fragments. So we can test if our model | + | fusion proteins of dCas9 and split-HRP fragments. So we can test if our model |
| − | reasonable by comparing the curves predicted by the model with those | + | is reasonable by comparing the curves predicted by the model with those |
| − | from the experiment. The results are shown below.</span> | + | obtained from the experiment. The results are shown below.</span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,801: | Line 1,832: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
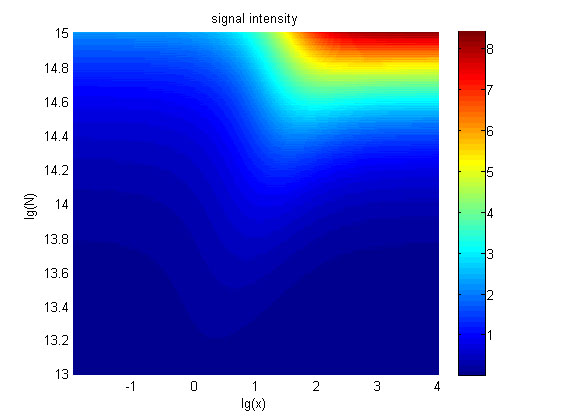
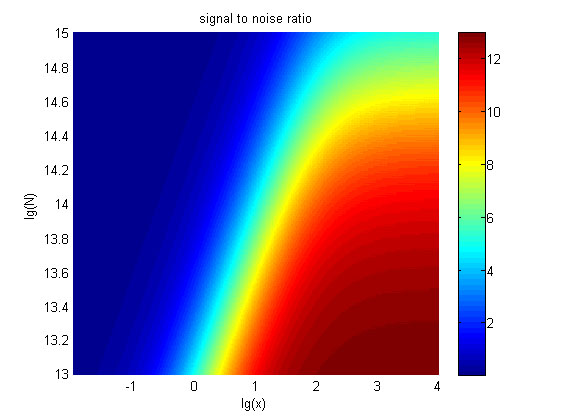
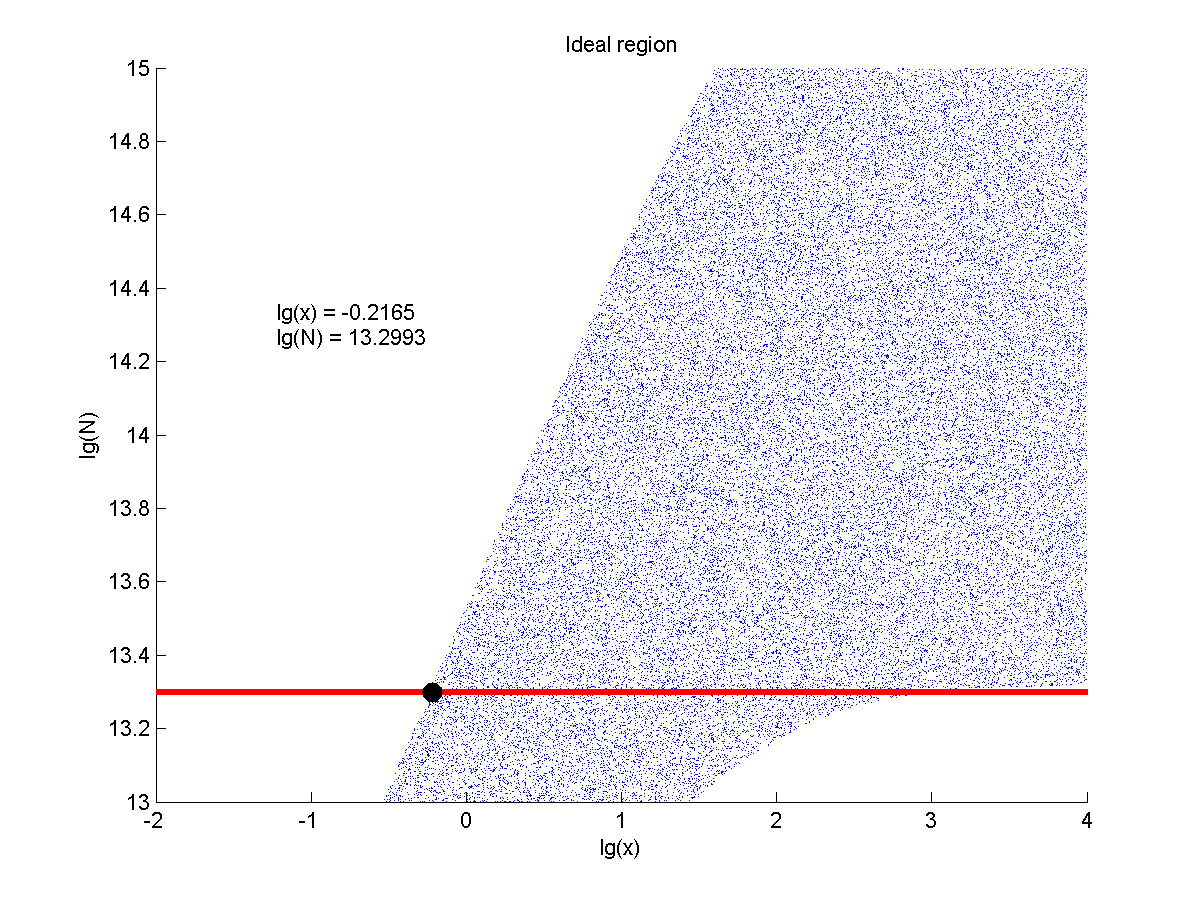
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Furthermore, | |
| − | + | we consider that it is conducive to signal detection when the value of signal | |
| − | + | intensity and SNR are both greater than two. We obtain the ideal region through | |
| − | + | calculation, which is shown as below. </span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,815: | Line 1,846: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA-- | + | [[File:T--NUDT_CHINA--modelfig7.jpg|700px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,837: | Line 1,868: | ||
the | the | ||
| − | value of <i>N</i> is set to e+13.3, the range of the value of<i> x</i> | + | value of <i>N</i> is set to e+13.3, the range of the value of<i> x</i> |
| − | in the ideal region is<span> maximum, which means when the value of the | + | contained in the ideal region is<span> maximum, which means when the value of |
| + | |||
| + | the | ||
molecule number of the fusion proteins is e+13.3, the concentration range of | molecule number of the fusion proteins is e+13.3, the concentration range of | ||
miRNA that can be detected is maximum. </span> | miRNA that can be detected is maximum. </span> | ||
| Line 1,909: | Line 1,942: | ||
size:18px;">1 Deng, R. <i>et al</i>. | size:18px;">1 Deng, R. <i>et al</i>. | ||
| − | Toehold-initiated rolling circle | + | Toehold-initiated rolling circle amplification for visualizing individual |
| − | amplification for visualizing individual | + | |
| − | <i>Angew Chem Int Ed Engl </i>53, 2389-2393, | + | microRNAs in situ in single cells. <i>Angew Chem Int Ed Engl </i>53, 2389- |
| − | doi:10.1002/anie.201309388 (2014).</span> | + | |
| + | 2393,doi:10.1002/anie.201309388 (2014).</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,955: | Line 1,988: | ||
.menu-link-container a:hover{ | .menu-link-container a:hover{ | ||
text-decoration:underline; | text-decoration:underline; | ||
| − | } #c_menu9909 .medium-6{ text-align:center; width:auto; border:solid | + | } #c_menu9909 .medium-6{ text-align:center; width:auto; border:solid |
| − | red; } #c_menu9909 .medium-6 h5{color:white;}#c_menu9909 .medium-6 | + | 0px red; } #c_menu9909 .medium-6 h5{color:white;}#c_menu9909 .medium-6 |
{height:315px;border:solid 0px red;}</style> | {height:315px;border:solid 0px red;}</style> | ||
| Line 2,005: | Line 2,038: | ||
<a href="/Team:NUDT_CHINA/Basic_Part">Basic Parts</a><br/> | <a href="/Team:NUDT_CHINA/Basic_Part">Basic Parts</a><br/> | ||
| − | <a href="/Team:NUDT_CHINA/Composite_Part">Composite Parts</a><br/> | + | <a href="/Team:NUDT_CHINA/Composite_Part">Composite |
| + | |||
| + | Parts</a><br/> | ||
<a href="/Team:NUDT_CHINA/Part_Collection">Part | <a href="/Team:NUDT_CHINA/Part_Collection">Part | ||
| Line 2,068: | Line 2,103: | ||
<img | <img | ||
| − | src="https://static.igem.org/mediawiki/2016/thumb/2/22/NUDT_CHINA2016_FILES-img-igem- | + | src="https://static.igem.org/mediawiki/2016/thumb/2/22/NUDT_CHINA2016_FILES-img- |
| + | |||
| + | igem-logo.png/800px-NUDT_CHINA2016_FILES-img-igem-logo.png" | ||
| − | + | style="width:300px;"> | |
<div class="content-open" style="width:300px;"> | <div class="content-open" style="width:300px;"> | ||
<p class="intro" style="font-size:18px;line-height:30px;"> | <p class="intro" style="font-size:18px;line-height:30px;"> | ||
| Line 2,224: | Line 2,261: | ||
var y = Math.max(y1, Math.max(y2, y3)); | var y = Math.max(y1, Math.max(y2, y3)); | ||
| − | // 滚动距离 = 目前距离 / 速度, 因为距离原来越小, 速度是大于 1 的数, | + | // 滚动距离 = 目前距离 / 速度, 因为距离原来越小, 速度是大于 1 的数, 所以滚动距 |
| − | + | 离会越来越小 | |
var speed = 1 + acceleration; | var speed = 1 + acceleration; | ||
window.scrollTo(Math.floor(x / speed), Math.floor(y / speed)); | window.scrollTo(Math.floor(x / speed), Math.floor(y / speed)); | ||
Revision as of 12:59, 18 October 2016