Qiuxinyuan12 (Talk | contribs) |
Qiuxinyuan12 (Talk | contribs) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | |||
<html> | <html> | ||
| Line 69: | Line 70: | ||
<meta name="HandheldFriendly" content="True" /> | <meta name="HandheldFriendly" content="True" /> | ||
<meta name="MobileOptimized" content="320" /> | <meta name="MobileOptimized" content="320" /> | ||
| − | + | <meta name="viewport" content="width=device-width, initial-scale=1.0",minimum-scale=0.2, maximum-scale=2.0, user-scalable=yes" /> | |
<meta content="on" http-equiv="cleartype" /> | <meta content="on" http-equiv="cleartype" /> | ||
<meta property="og:title" content="Home" /> | <meta property="og:title" content="Home" /> | ||
| Line 88: | Line 89: | ||
<div class="inner-wrap"> | <div class="inner-wrap"> | ||
<!-- Off Canvas Menu --> | <!-- Off Canvas Menu --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- close the off-canvas menu --> | <!-- close the off-canvas menu --> | ||
<a class="exit-off-canvas"></a> | <a class="exit-off-canvas"></a> | ||
| Line 152: | Line 116: | ||
<a class="nav-global__link" title="" href="#">AWARDS</a></li> | <a class="nav-global__link" title="" href="#">AWARDS</a></li> | ||
</ul> | </ul> | ||
| − | <style>#menu06{width:200px;right:0px;} #menu07{width:200px;right:0px;} | + | <style> |
| + | .content{ | ||
| + | box-shadow: 6px 6px 3px #888888; | ||
| + | } | ||
| + | #menu06{width:200px;right:0px;} #menu07{width:200px;right:0px;} | ||
#menu01 .arrow-up{margin-right:6.429rem;} | #menu01 .arrow-up{margin-right:6.429rem;} | ||
#menu02 .arrow-up{margin-right:90px;} #menu03 .arrow-up{margin-right:90px;} #menu06 .arrow-up{margin-right:120px;} #menu07 .arrow-up{margin-right:27px;} #menu01{width:200px;right:31rem;} #menu02{width:200px;right:27rem;} #menu03{width:200px;right:21.3rem;} | #menu02 .arrow-up{margin-right:90px;} #menu03 .arrow-up{margin-right:90px;} #menu06 .arrow-up{margin-right:120px;} #menu07 .arrow-up{margin-right:27px;} #menu01{width:200px;right:31rem;} #menu02{width:200px;right:27rem;} #menu03{width:200px;right:21.3rem;} | ||
| Line 322: | Line 290: | ||
</div> | </div> | ||
</nav> | </nav> | ||
| − | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } }</style> | + | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } } |
| + | |||
| + | #feature{ | ||
| + | background-size:100%; | ||
| + | } | ||
| + | @media screen and (max-width: 1200px) { | ||
| + | #feature{ | ||
| + | background-size:1200px; | ||
| + | } | ||
| + | } | ||
| + | </style> | ||
<div class="breadcrumb-share-wrap"> | <div class="breadcrumb-share-wrap"> | ||
| Line 335: | Line 313: | ||
<!-- end .row --></div> | <!-- end .row --></div> | ||
<!-- end #header-outer-wrap--> | <!-- end #header-outer-wrap--> | ||
| − | <div id="feature" class="row collapse text-light feature-footer-active"> | + | <div id="feature" class="row collapse text-light feature-footer-active" style="background-color: rgb(3,13,38);background-image:url(https://static.igem.org/mediawiki/2016/thumb/3/3f/NUDT_CHINA2016_FILES-img-banner-3.0.png/800px-NUDT_CHINA2016_FILES-img-banner-3.0.png);background-position:0px -350px;background-repeat:no-repeat;"> |
<div class="inner-wrap medium-24 columns"> | <div class="inner-wrap medium-24 columns"> | ||
<!--TYPO3SEARCH_begin--> | <!--TYPO3SEARCH_begin--> | ||
| Line 341: | Line 319: | ||
<div class="carousel " id="slideshow-4811" data-cycle-log="false" data-cycle-auto-height="calc" data-cycle-prev="#prev-4811" data-cycle-next="#next-4811" data-cycle-pager="#cycle-pager-4811" data-cycle-easing="easeInOutQuad" data-cycle-fx="scrollVertUp" data-cycle-center-horz="true" data-cycle-speed="1000" data-cycle-timeout="8000" data-cycle-paused="true" data-cycle-slides="> div.slide"> | <div class="carousel " id="slideshow-4811" data-cycle-log="false" data-cycle-auto-height="calc" data-cycle-prev="#prev-4811" data-cycle-next="#next-4811" data-cycle-pager="#cycle-pager-4811" data-cycle-easing="easeInOutQuad" data-cycle-fx="scrollVertUp" data-cycle-center-horz="true" data-cycle-speed="1000" data-cycle-timeout="8000" data-cycle-paused="true" data-cycle-slides="> div.slide"> | ||
<div class="slide first"> | <div class="slide first"> | ||
| − | <link rel="stylesheet" href="/Team | + | <link rel="stylesheet" href="/Team--NUDT_CHINA/CSS3.css" /> |
<!-- Quick initialize with theme color from first slide --> | <!-- Quick initialize with theme color from first slide --> | ||
<div class="row collapse " id="c6601" data-theme-class="bh-color__theme-c6601"> | <div class="row collapse " id="c6601" data-theme-class="bh-color__theme-c6601"> | ||
<div class="columns"> | <div class="columns"> | ||
| − | <section class="big-header bh__height-size-3 text-light bh__video"> | + | <section class="big-header bh__height-size-3 text-light bh__video" style="height:300px;"> |
| − | <div class="bh__background bh__background-color | + | <div class="bh__background bh__background-color "></div> |
| − | <div class="bh__content-wrap"> | + | <div class="bh__content-wrap" style="height:120px;"> |
<div class="bh__content "> | <div class="bh__content "> | ||
<div class="bh__content-inner-wrap"> | <div class="bh__content-inner-wrap"> | ||
<!--h3 class="bh__subtitle"><a href="http#/explore/whats-here/exhibits/brick-by-brick/"><span class="line-wrap"><span class="line"><span class="bh__info">Exhibit / </span>Brick by Brick</span></span></a></h3--> | <!--h3 class="bh__subtitle"><a href="http#/explore/whats-here/exhibits/brick-by-brick/"><span class="line-wrap"><span class="line"><span class="bh__info">Exhibit / </span>Brick by Brick</span></span></a></h3--> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<h2 style="font-size:36px;" class="bh__title "> | <h2 style="font-size:36px;" class="bh__title "> | ||
<!--<a href="#">--> | <!--<a href="#">--> | ||
| − | + | <span style="line-height:46px;" class="line-wrap"> | |
| − | + | <span style="line-height:46px;" class="line">Development of A Novel</span></span> | |
| − | + | <span style="line-height:46px;" class="line-wrap"> | |
| − | + | <span style="line-height:46px;" class="line">Blood-MicroRNA Handy Detection System with CRISPR</span></span> | |
| − | <!--</a>--> | + | <!--</a>--></h2> |
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 378: | Line 346: | ||
<div class="cycle-pager" id="cycle-pager-4811"></div> | <div class="cycle-pager" id="cycle-pager-4811"></div> | ||
</div> | </div> | ||
| − | + | </div> | |
| − | + | </div> | |
<!-- end #feature --> | <!-- end #feature --> | ||
<!-- end .columns --></div> | <!-- end .columns --></div> | ||
| − | <div style="margin-top: | + | <div style="margin-top:20px;" id="content-wrap-ink"><!-- adjust in add_JS --> |
| − | <div class="row footer-link" style=""> | + | <div class="row footer-link" style=""> |
| + | <div style="text-align:right;"><h5> | ||
| + | <a style="color:rgb(10,31,84);" href="/Team:NUDT_CHINA">HomePage</a> • | ||
| + | <!-- 修改这里!! -->PROJECT • | ||
| + | <a style="color:rgb(10,31,84);" href="/Team:NUDT_CHINA/Design"><!-- 修改这里!! -->Design</a> | ||
| + | </h5><hr style="width:40%;margin-left:60%;border-top:1px solid rgb(10,31,84);" /></div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h1><span style="color:#7f1015">Design</span></h1> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">General Design</span></span><hr /> | ||
| + | </h2> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To develop a novel cell-free platform for | ||
| + | the low-cost, high-efficient and visualized detection of serum miRNAs, two | ||
| + | essential systems, namely RCA based DNA amplification system, and dCas9 | ||
| + | conjugated split-reporting system, were combined, modified, and then assessed | ||
| + | in our project.</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The first system, RCA based DNA | ||
| + | amplification system, was introduced to primarily amplify the input miRNA | ||
| + | signal with a high specificity under an isothermal and moderate condition. | ||
| + | Specifically, a dumbbell shaped probe containing a tunable toehold domain on | ||
| + | its loop was custom designed and prepared for the detection of a specific | ||
| + | target miRNA. Target miRNAs could bind with the toehold domain, and then | ||
| + | trigger the toehold-mediated strand displacement (TMSD) process resulting in a | ||
| + | switch of the probe from the dumbbell shaped form into a circular form (termed | ||
| + | as initiated probe, or iprobe in brief). The iprobe could then be used as the | ||
| + | template for the subsequent RCA reaction (Figure 1, left panel). A mismatched | ||
| + | miRNA, however, would fail to trigger the TMSD due to the resistance of the | ||
| + | stabilized dumbbell structure, thus producing no amplification products. Once | ||
| + | RCA products were produced, a traditional Sybr I based fluorescence assay could | ||
| + | be conducted to assess the effectiveness of RCA based DNA amplification system. | ||
| + | A fluoresce microplate reader would be needed for such matters (Figure 1, | ||
| + | middle panel). </span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT CHINA--designfig1.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
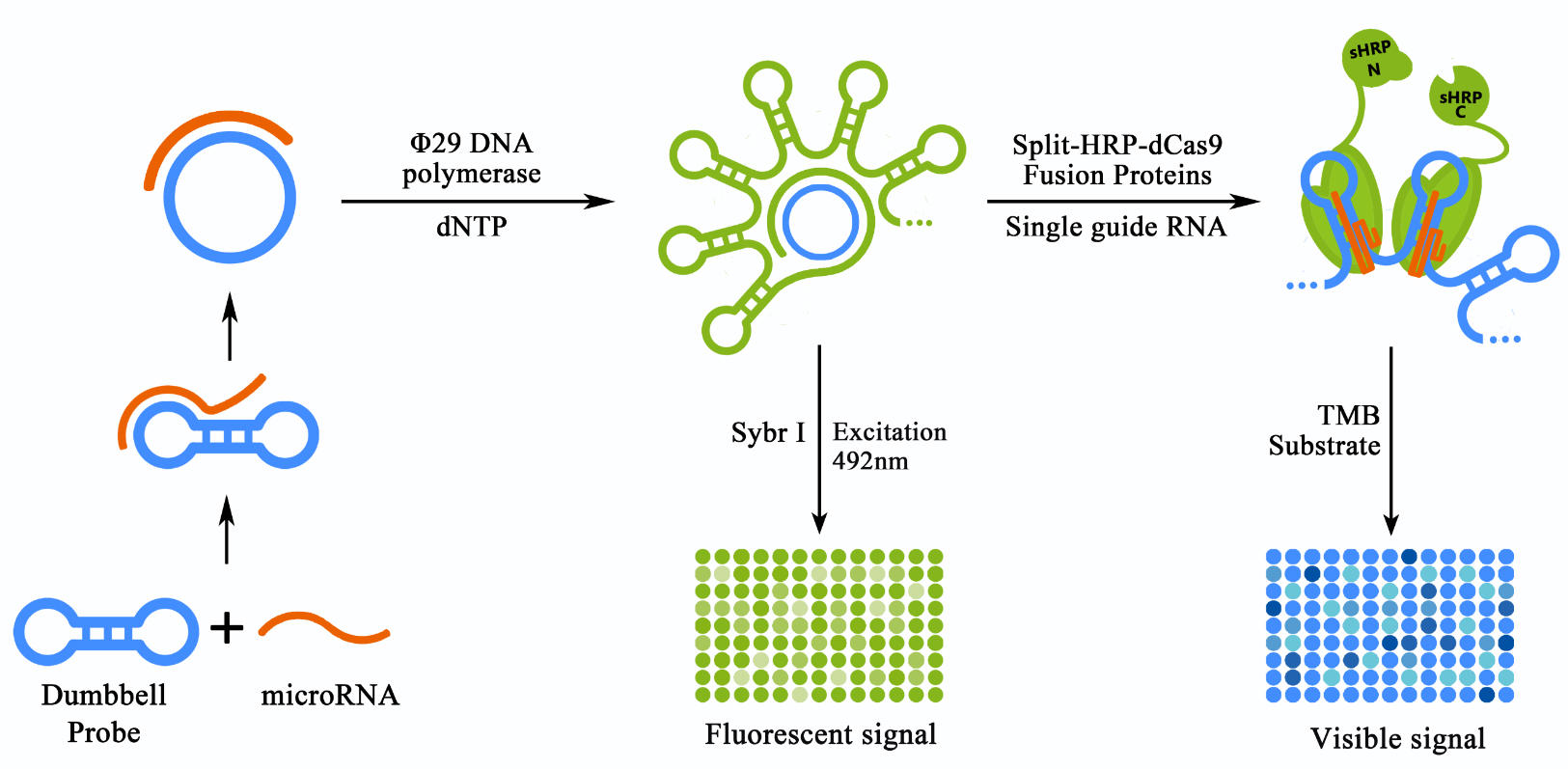
| + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Figure 1. Schematic representation of the detection process of our scheme.</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">The miRNA in the sample can bind with the pre-designed dumbbell probe. Once bond, the chain-replace reaction would be activated and the dumbbell shaped probe would be transformed into a circular structure, thus allowed the initiation of the rolling cycle DNA amplification (RCA) process. The product of RCA reaction can be detected directly through Sybr I staining and a fluorescent plate reader. The RCA product were to be bound by sgRNA guided dCas9 proteins fused with split-HRP reporters, thus produced a visible and stronger signal by producing TMB diamine from TMB substrate.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <p align="center" style="text-align:center;text-indent:22.1pt;"> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span></b> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In order to achieve the further | ||
| + | amplification and visualization of the RCA output signal, the dCas9 conjugated | ||
| + | split-reporting system was then introduced into our scheme. The fusion proteins | ||
| + | of dCas9 and split-HRP fragments, namely sHRP-N-dCas9 (N-sHdC) and sHRP-C-dCas9 | ||
| + | (C-sHdC) could be obtained from genetically modified </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">E. coli</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> strains containing relevant expression plasmids. With the | ||
| + | guidance of sgRNA, N-sHdC proteins and C-sHdC proteins would be able to bind | ||
| + | randomly to the numerous double-strand loci on the RCA product (Figure 2, right | ||
| + | panel). For all those bound to loci that were close enough, split-HRP fragments | ||
| + | would interact with each other and HRP enzyme activity would be retained. | ||
| + | Building on this, HRP enzyme activity could then be determined by adding | ||
| + | substrates such as 3,3',5,5'-Tetramethylbenzidine (TMB) with a visual output | ||
| + | signal.</span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT CHINA--designfig2.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
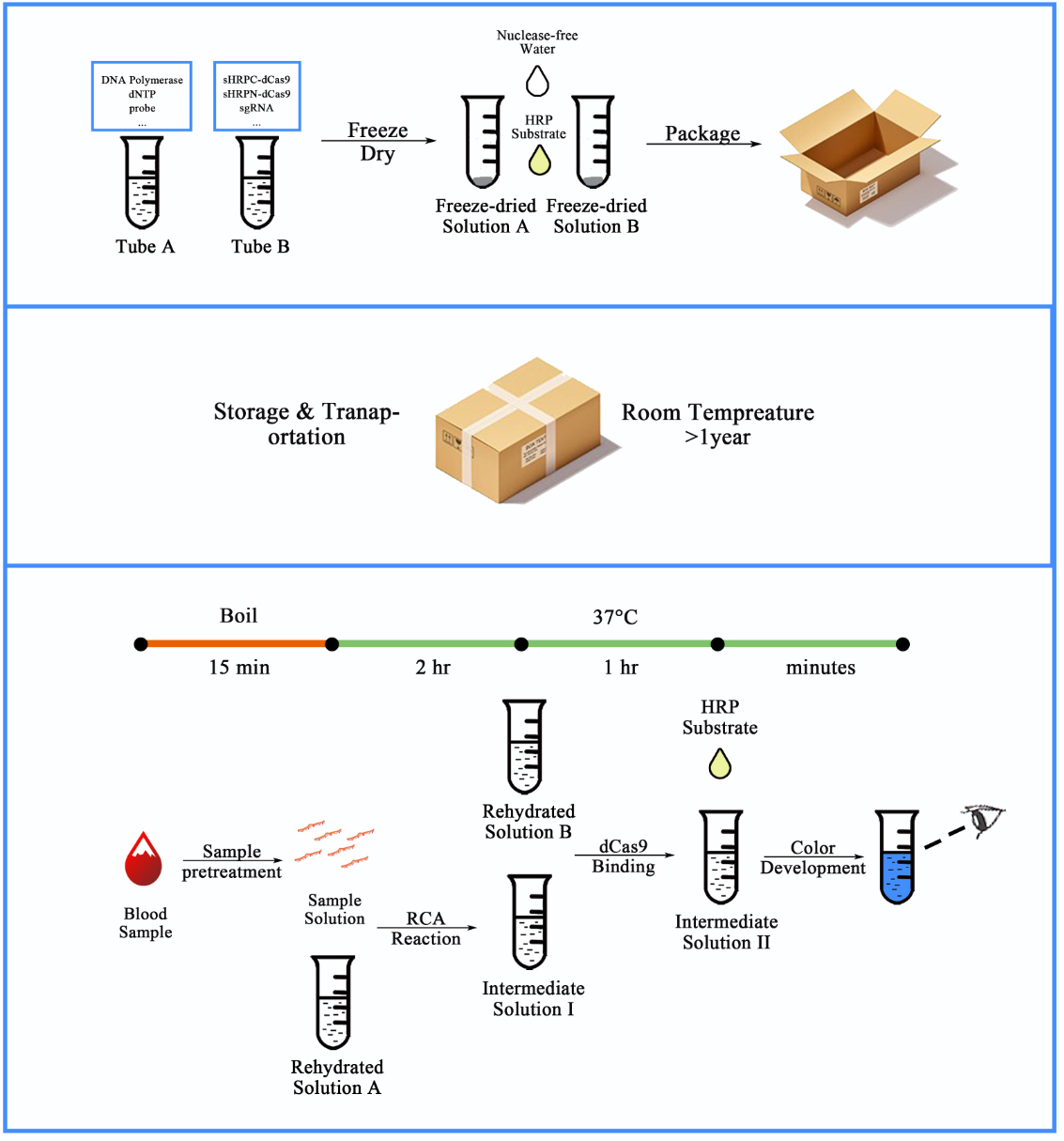
| + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Figure 2. Workflow for our CRISPR-based blood-microRNA detection system.</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">The synthesized and sealed dumbbell probe together with other RCA related materials can be embedded into tube A, and purified fusion proteins of dCas9 and split-HRP fragments together with other dCas9-binding related materials can be embedded into tube B. The reagents in both tubes can be freeze-dried to maintain stable in room temperature for a relatively long time (>1 year). When entering the detection process, serum samples were pre-treated by boiling for 15 min to expose the miRNAs from molecule complexes and exosomes. After a series of simple operations, the amount of the specific RNA could be indicated by the color depth from the colorless to the dark blue. | ||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p align="center" style="text-align:center;text-indent:22.1pt;"> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span></b> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Since most proteins could remain stable | ||
| + | under normal storage conditions if freeze dried, and retain their activity | ||
| + | after rehydrated. Our system could then be developed into a kit that contains | ||
| + | freeze-dried components together with other liquid components that is stable in | ||
| + | regular storage conditions, such as TMB substrate and DEPC treated water (Figure | ||
| + | 2, upper and middle panel). Meanwhile, with its advantage in visualized outputs | ||
| + | and moderate temperature (37</span><span style="line-height:2;font-family:Perpetua;font-size:18px;">℃</span><span style="line-height:2;font-family:Perpetua;font-size:18px;">) detection process, such | ||
| + | kit would be able to be deployed in low-resource settings and dramatically | ||
| + | lower the cost of and technical barrier for wider cancer scanning and early | ||
| + | detection.</span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Prototype: serum let-7a detection system</span></span><hr /> | ||
| + | </h2> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To validate and demonstrate our design, miR | ||
| + | let-7a, as an important serum biomarker for non-small cell lung cancer, was | ||
| + | chosen as the target miRNA. Previously, miR let-7a has been reported to be | ||
| + | down-regulated for 20%-40% in serum samples from NSCLC patients compared to | ||
| + | healthy people </span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">1</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">.</span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To produce miR let-7a for further detection, | ||
| + | we put the sequence of let-7a under control of a T7 promoter. The plasmid was | ||
| + | linearized at the point right after let-7a sequence and </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">in vitro</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> transcription kit was then used for the transcription of | ||
| + | let-7a.</span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">AS A PROOF OF CONCEPT, let-7a was diluted in | ||
| + | DEPC-treated water on various concentrations to assess the reliability, sensibility | ||
| + | and specificity of our scheme. To begin with, four different probes were | ||
| + | designed to be probe candidates for the RCA reaction based on let7a sequence | ||
| + | and probe design principles. Once they have been synthesized and purified, RCA | ||
| + | reactions with 10nM let-7a input were performed against all four probes to | ||
| + | select the optimal probe for further test. By using this probe, the sensibility | ||
| + | and specificity of RCA reaction were then determined with sybr I fluorescence | ||
| + | assay. Moreover, N-sHdC and C-sHdC protein were expressed and purified from </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">E.coli</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;">, and subsequently used for the | ||
| + | dCas9 binding process together with an </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">in | ||
| + | vitro</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> expressed sgRNA. TMB substrate was used to test the HRP activity. | ||
| + | (See proof of concept page for more details)</span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">FOR FURTHER DEMONSTRATION OF OUR PROJECT, let-7a | ||
| + | was dissolved in 7% mixed human serum collected from 50 healthy volunteers. | ||
| + | Similarly, sybr I fluorescence assay and HRP activity assay was conducted to | ||
| + | verify the reliability, sensibility and specificity of our scheme in stimulated | ||
| + | clinical samples. MOREOVER, serum samples collected from NSCLC patients and | ||
| + | healthy volunteers were also tested after different pretreatment for further | ||
| + | demonstration of our scheme. (See demonstration page for more details)</span> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Reference</span></span><hr /> | ||
| + | </h2> | ||
| + | <p style="text-indent:-0.55pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Jeong, H. C.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> Aberrant expression of let-7a miRNA in the blood of | ||
| + | non-small cell lung cancer patients. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Mol | ||
| + | Med Rep</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">4</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 383-387, | ||
| + | doi:10.3892/mmr.2011.430 (2011).</span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 392: | Line 555: | ||
| − | |||
| − | |||
| Line 406: | Line 567: | ||
</div> | </div> | ||
<!-- end #content-wrap --> | <!-- end #content-wrap --> | ||
| − | + | <div style="border:solid 1px rgb(10,31,84);background-color:rgb(10,31,84);"> | |
| − | + | <div class="row footer-link" style="border:solid 0px red;margin-top:100px;"> | |
| − | + | <style>.menu-link-container ul li{ list-style:none; } .menu-link-container a{ color:white; font-size:14px;line-height:26px; } | |
| − | + | .menu-link-container a:hover{ | |
| − | + | text-decoration:underline; | |
| − | + | } #c_menu9909 .medium-6{ text-align:center; width:auto; border:solid 0px red; } #c_menu9909 .medium-6 h5{color:white;}#c_menu9909 .medium-6{height:315px;border:solid 0px red;}</style> | |
| − | + | <div id="c_menu9909"> | |
| − | + | <!--div class="medium-6 columns" style="width:auto;"> | |
| − | + | <h5 class="column-header">HOME</h5> | |
| − | + | <div class="menu-link-container"></div></div--> | |
| − | + | <div class="medium-6 columns" style="width:auto;"> | |
| − | + | <h5 class="column-header">TEAM</h5> | |
| − | + | <div class="menu-link-container"> | |
| − | + | ||
| − | + | ||
| − | + | <a href="/Team:NUDT_CHINA/Team">Team</a><br/> | |
| − | + | ||
| − | + | <a href="/Team:NUDT_CHINA/Collaborations">Collaborations</a><br/> | |
| − | + | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">PROJECT</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Description">Description</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Design">Design</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Experiments">Experiments</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Proof">Proof of Concept</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Demonstrate">Demonstrate</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Results">Results</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Notebook">Notebook</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">PARTS</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Parts">Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Basic_Part">Basic Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Composite_Part">Composite Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Part_Collection">Part Collection</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <a href="/Team:NUDT_CHINA/Safety" style="text-decoration:none;"> | ||
| + | <h5 class="column-header">SAFETY</h5></a> | ||
| + | |||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <a href="/Team:NUDT_CHINA/Attributions" style="text-decoration:none;"> | ||
| + | <h5 class="column-header">ATTRIBUTIONS</h5></a> | ||
| + | |||
| + | </div> | ||
| + | <!--div class="medium-6 columns" style="width:auto;"> | ||
| + | <A href="/Team:NUDT_CHINA/Attributions" style="text-decoration:none;"><h5 class="column-header">ATTRIBUTIONS</h5></A> | ||
| + | <div class="menu-link-container"></div></div--> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">HUMAN | ||
| + | <br>PRACTICES</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Human_Practices">Human Practices</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/HP/Silver">Silver</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/HP/Gold">Gold</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Integrated_Practices">Integrated Practices</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Engagement">Engagement</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">AWARDS</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Model">Model</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:0px;height:0px;"></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
<footer id="footer"> | <footer id="footer"> | ||
<div class="row footer-link"> | <div class="row footer-link"> | ||
| Line 556: | Line 718: | ||
--> | --> | ||
| − | + | <!-- to top edit by inksci---> | |
| + | <style> | ||
| + | #to_top{ | ||
| + | position:fixed; | ||
| + | bottom:100px; | ||
| + | right:0px; | ||
| + | width:60px; | ||
| + | height:60px; | ||
| + | font-size:14px; | ||
| + | line-height:60px; | ||
| + | text-align:center; | ||
| + | background-color:black; | ||
| + | color:rgb(246,168,0); | ||
| + | cursor:pointer; | ||
| + | } | ||
| + | </style> | ||
| + | <div onclick="top_it()" id="to_top"> | ||
| + | TOP | ||
| + | </div> | ||
| + | <script type="text/javascript"> | ||
| + | var oTop = document.getElementById("to_top"); | ||
| + | oTop.style.display="none"; | ||
| + | var top_break=false; | ||
| + | /** | ||
| + | * 回到页面顶部 | ||
| + | * @param acceleration 加速度 | ||
| + | * @param time 时间间隔 (毫秒) | ||
| + | **/ | ||
| + | var save_scrolltop=100000000000000; | ||
| + | window.onscroll = function(){ | ||
| + | |||
| + | |||
| + | var scrolltop = document.documentElement.scrollTop || document.body.scrollTop; | ||
| + | if(scrolltop>300){ | ||
| + | oTop.style.display="block"; | ||
| + | }else{ | ||
| + | oTop.style.display="none"; | ||
| + | } | ||
| + | if(scrolltop>save_scrolltop) | ||
| + | { | ||
| + | top_break=true; | ||
| + | } | ||
| + | save_scrolltop=scrolltop; | ||
| + | } | ||
| + | function top_it(){ | ||
| + | top_break=false; | ||
| + | goTop();return false; | ||
| + | } | ||
| + | function goTop(acceleration, time) { | ||
| + | if(top_break){ | ||
| + | return; | ||
| + | } | ||
| + | oTop.style.display="none"; | ||
| + | acceleration = acceleration || 0.1; | ||
| + | time = time || 16; | ||
| + | |||
| + | var x1 = 0; | ||
| + | var y1 = 0; | ||
| + | var x2 = 0; | ||
| + | var y2 = 0; | ||
| + | var x3 = 0; | ||
| + | var y3 = 0; | ||
| + | |||
| + | if (document.documentElement) { | ||
| + | x1 = document.documentElement.scrollLeft || 0; | ||
| + | y1 = document.documentElement.scrollTop || 0; | ||
| + | } | ||
| + | if (document.body) { | ||
| + | x2 = document.body.scrollLeft || 0; | ||
| + | y2 = document.body.scrollTop || 0; | ||
| + | } | ||
| + | var x3 = window.scrollX || 0; | ||
| + | var y3 = window.scrollY || 0; | ||
| + | |||
| + | // 滚动条到页面顶部的水平距离 | ||
| + | var x = Math.max(x1, Math.max(x2, x3)); | ||
| + | // 滚动条到页面顶部的垂直距离 | ||
| + | var y = Math.max(y1, Math.max(y2, y3)); | ||
| + | |||
| + | // 滚动距离 = 目前距离 / 速度, 因为距离原来越小, 速度是大于 1 的数, 所以滚动距离会越来越小 | ||
| + | var speed = 1 + acceleration; | ||
| + | window.scrollTo(Math.floor(x / speed), Math.floor(y / speed)); | ||
| + | |||
| + | // 如果距离不为零, 继续调用迭代本函数 | ||
| + | if(x > 0 || y > 0) { | ||
| + | var invokeFunction = "goTop(" + acceleration + ", " + time + ")"; | ||
| + | window.setTimeout(invokeFunction, time); | ||
| + | } | ||
| + | } | ||
| + | </script> | ||
| + | <!-- to top edit by inksci---> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 01:47, 19 October 2016
TOP