Qiuxinyuan12 (Talk | contribs) |
Qiuxinyuan12 (Talk | contribs) |
||
| Line 365: | Line 365: | ||
| + | <p> | ||
| + | <h1> | ||
| + | <span><span style="color:#7f1015">Team Parts</span></span><hr /> | ||
| + | </h1> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This year, we handed in 26 high quality, well documented bio-brick | ||
| + | parts, including all those we used in our project this year, and several | ||
| + | brilliantly designed others forming a Protein-protein Interaction toolkit (PPI | ||
| + | toolkit) inspired by our iGEM project within these two years.</span> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | |||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Favorite Parts</span></span><hr /> | ||
| + | </h2> | ||
| + | <p> | ||
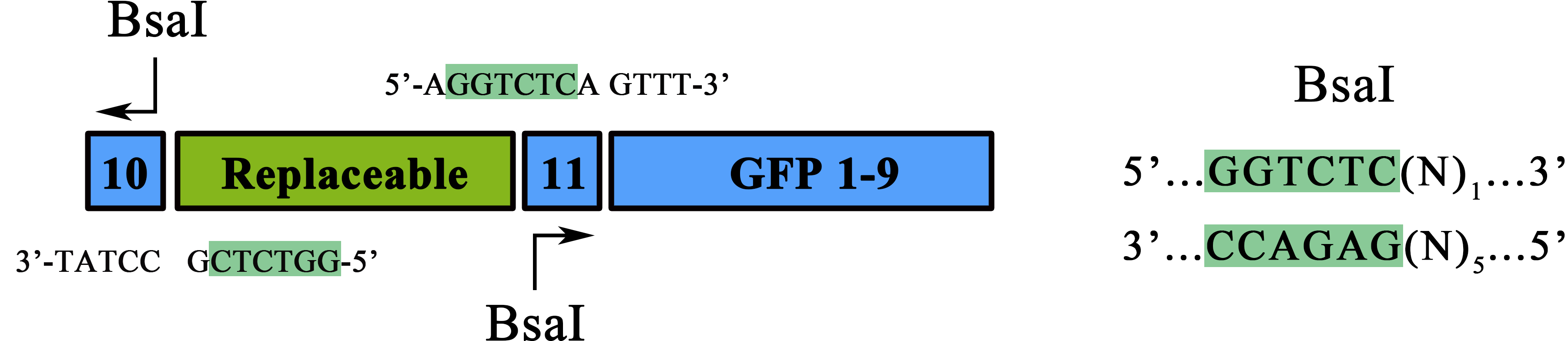
| + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">PPI | ||
| + | reporter based on split-GFP</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">(BBa_K1997014)</span> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--partsfig1.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This part is a split-GFP reporter used for PPI study. Other than the | ||
| + | traditional N-GFP, C-GFP approach, another split method on GFP was introduced | ||
| + | to reach better reads and higher NSR. Special designs were taken for to | ||
| + | optimize the applicability and adaptive of such parts. Specifically, a novel | ||
| + | designed substitution method allowed a one-step replacement of the | ||
| + | “Replaceable” marked region to be replaced by a “Protein1-RBS-Protein2” form | ||
| + | fragment for studying the interaction between protein1 and protein2, thus | ||
| + | dramatically simplified the cloning process.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | <p> | ||
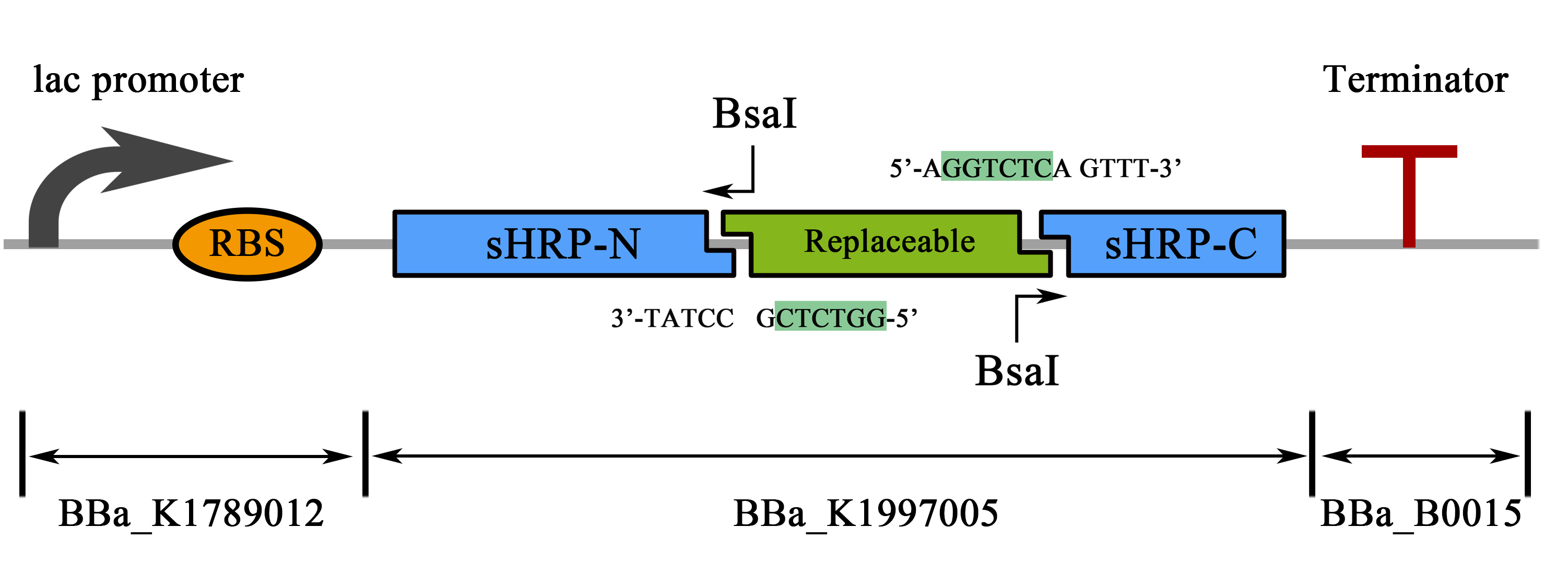
| + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Split-HRP | ||
| + | based PPI Detection device</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">(BBa_K1997011)</span> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--partsfig2.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| Line 376: | Line 452: | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This part is a composite part containing a lac promoter, a split-HRP | ||
| + | reporter and a double terminator designed for the usage of PPI study. Similarly, | ||
| + | an implanted novel substitution method allowed a one-step replacement of the | ||
| + | “Replaceable” marked region to be replaced by a “Protein 1-RBS-Protein 2” form | ||
| + | fragment for studying the interaction between protein 1 and protein 2, thus | ||
| + | dramatically simplified the cloning process.</span> | ||
| + | </p> | ||
| + | </p> | ||
| + | <p> | ||
| + | <br /> | ||
| + | </p> | ||
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Parts List</span></span><hr /> | ||
| + | </h2> | ||
<center> | <center> | ||
</html> | </html> | ||
Revision as of 03:50, 19 October 2016
TOP