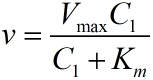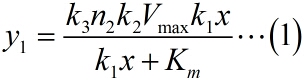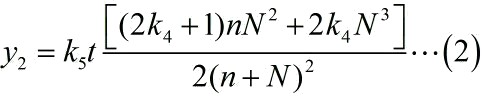Zhuchushu13 (Talk | contribs) |
Zhuchushu13 (Talk | contribs) |
||
| (43 intermediate revisions by 2 users not shown) | |||
| Line 69: | Line 69: | ||
<meta name="HandheldFriendly" content="True" /> | <meta name="HandheldFriendly" content="True" /> | ||
<meta name="MobileOptimized" content="320" /> | <meta name="MobileOptimized" content="320" /> | ||
| − | + | <meta name="viewport" content="width=device-width, initial-scale=1.0",minimum-scale=0.2, maximum-scale=2.0, user-scalable=yes" /> | |
<meta content="on" http-equiv="cleartype" /> | <meta content="on" http-equiv="cleartype" /> | ||
<meta property="og:title" content="Home" /> | <meta property="og:title" content="Home" /> | ||
| Line 88: | Line 88: | ||
<div class="inner-wrap"> | <div class="inner-wrap"> | ||
<!-- Off Canvas Menu --> | <!-- Off Canvas Menu --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- close the off-canvas menu --> | <!-- close the off-canvas menu --> | ||
<a class="exit-off-canvas"></a> | <a class="exit-off-canvas"></a> | ||
| Line 152: | Line 115: | ||
<a class="nav-global__link" title="" href="#">AWARDS</a></li> | <a class="nav-global__link" title="" href="#">AWARDS</a></li> | ||
</ul> | </ul> | ||
| − | <style>#menu06{width:200px;right:0px;} #menu07{width:200px;right:0px;} | + | <style> |
| + | .content{ | ||
| + | box-shadow: 6px 6px 3px #888888; | ||
| + | } | ||
| + | #menu06{width:200px;right:0px;} #menu07{width:200px;right:0px;} | ||
#menu01 .arrow-up{margin-right:6.429rem;} | #menu01 .arrow-up{margin-right:6.429rem;} | ||
#menu02 .arrow-up{margin-right:90px;} #menu03 .arrow-up{margin-right:90px;} #menu06 .arrow-up{margin-right:120px;} #menu07 .arrow-up{margin-right:27px;} #menu01{width:200px;right:31rem;} #menu02{width:200px;right:27rem;} #menu03{width:200px;right:21.3rem;} | #menu02 .arrow-up{margin-right:90px;} #menu03 .arrow-up{margin-right:90px;} #menu06 .arrow-up{margin-right:120px;} #menu07 .arrow-up{margin-right:27px;} #menu01{width:200px;right:31rem;} #menu02{width:200px;right:27rem;} #menu03{width:200px;right:21.3rem;} | ||
| Line 322: | Line 289: | ||
</div> | </div> | ||
</nav> | </nav> | ||
| − | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } }</style> | + | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } } |
| + | |||
| + | #feature{ | ||
| + | background-size:100%; | ||
| + | } | ||
| + | @media screen and (max-width: 1200px) { | ||
| + | #feature{ | ||
| + | background-size:1200px; | ||
| + | } | ||
| + | } | ||
| + | </style> | ||
<div class="breadcrumb-share-wrap"> | <div class="breadcrumb-share-wrap"> | ||
| Line 335: | Line 312: | ||
<!-- end .row --></div> | <!-- end .row --></div> | ||
<!-- end #header-outer-wrap--> | <!-- end #header-outer-wrap--> | ||
| − | <div id="feature" class="row collapse text-light feature-footer-active"> | + | <div id="feature" class="row collapse text-light feature-footer-active" style="background-color: rgb(3,13,38);background-image:url(https://static.igem.org/mediawiki/2016/thumb/3/3f/NUDT_CHINA2016_FILES-img-banner-3.0.png/800px-NUDT_CHINA2016_FILES-img-banner-3.0.png);background-position:0px -350px;background-repeat:no-repeat;"> |
<div class="inner-wrap medium-24 columns"> | <div class="inner-wrap medium-24 columns"> | ||
<!--TYPO3SEARCH_begin--> | <!--TYPO3SEARCH_begin--> | ||
| Line 341: | Line 318: | ||
<div class="carousel " id="slideshow-4811" data-cycle-log="false" data-cycle-auto-height="calc" data-cycle-prev="#prev-4811" data-cycle-next="#next-4811" data-cycle-pager="#cycle-pager-4811" data-cycle-easing="easeInOutQuad" data-cycle-fx="scrollVertUp" data-cycle-center-horz="true" data-cycle-speed="1000" data-cycle-timeout="8000" data-cycle-paused="true" data-cycle-slides="> div.slide"> | <div class="carousel " id="slideshow-4811" data-cycle-log="false" data-cycle-auto-height="calc" data-cycle-prev="#prev-4811" data-cycle-next="#next-4811" data-cycle-pager="#cycle-pager-4811" data-cycle-easing="easeInOutQuad" data-cycle-fx="scrollVertUp" data-cycle-center-horz="true" data-cycle-speed="1000" data-cycle-timeout="8000" data-cycle-paused="true" data-cycle-slides="> div.slide"> | ||
<div class="slide first"> | <div class="slide first"> | ||
| − | <link rel="stylesheet" href="/Team | + | <link rel="stylesheet" href="/Team--NUDT_CHINA/CSS3.css" /> |
<!-- Quick initialize with theme color from first slide --> | <!-- Quick initialize with theme color from first slide --> | ||
<div class="row collapse " id="c6601" data-theme-class="bh-color__theme-c6601"> | <div class="row collapse " id="c6601" data-theme-class="bh-color__theme-c6601"> | ||
<div class="columns"> | <div class="columns"> | ||
| − | <section class="big-header bh__height-size-3 text-light bh__video"> | + | <section class="big-header bh__height-size-3 text-light bh__video" style="height:300px;"> |
| − | <div class="bh__background bh__background-color | + | <div class="bh__background bh__background-color "></div> |
| − | <div class="bh__content-wrap"> | + | <div class="bh__content-wrap" style="height:120px;"> |
<div class="bh__content "> | <div class="bh__content "> | ||
<div class="bh__content-inner-wrap"> | <div class="bh__content-inner-wrap"> | ||
<!--h3 class="bh__subtitle"><a href="http#/explore/whats-here/exhibits/brick-by-brick/"><span class="line-wrap"><span class="line"><span class="bh__info">Exhibit / </span>Brick by Brick</span></span></a></h3--> | <!--h3 class="bh__subtitle"><a href="http#/explore/whats-here/exhibits/brick-by-brick/"><span class="line-wrap"><span class="line"><span class="bh__info">Exhibit / </span>Brick by Brick</span></span></a></h3--> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<h2 style="font-size:36px;" class="bh__title "> | <h2 style="font-size:36px;" class="bh__title "> | ||
<!--<a href="#">--> | <!--<a href="#">--> | ||
| − | + | <span style="line-height:46px;" class="line-wrap"> | |
| − | + | <span style="line-height:46px;" class="line">Development of A Novel</span></span> | |
| − | + | <span style="line-height:46px;" class="line-wrap"> | |
| − | + | <span style="line-height:46px;" class="line">Blood-MicroRNA Handy Detection System with CRISPR</span></span> | |
| − | <!--</a>--> | + | <!--</a>--></h2> |
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 378: | Line 345: | ||
<div class="cycle-pager" id="cycle-pager-4811"></div> | <div class="cycle-pager" id="cycle-pager-4811"></div> | ||
</div> | </div> | ||
| − | + | </div> | |
| − | + | </div> | |
<!-- end #feature --> | <!-- end #feature --> | ||
<!-- end .columns --></div> | <!-- end .columns --></div> | ||
| − | <div style="margin-top: | + | <div style="margin-top:20px;" id="content-wrap-ink"><!-- adjust in add_JS --> |
| − | <div class="row footer-link" style=""> | + | <div class="row footer-link" style=""> |
| + | <div style="text-align:right;"><h5> | ||
| + | <a style="color:rgb(10,31,84);" href="/Team:NUDT_CHINA">HomePage</a> • | ||
| + | <!-- 修改这里!! -->AWARDS • | ||
| + | <a style="color:rgb(10,31,84);" href="/Team:NUDT_CHINA/Model"><!-- 修改这里!! -->Model</a> | ||
| + | </h5><hr style="width:40%;margin-left:60%;border-top:1px solid rgb(10,31,84);" /></div> | ||
| Line 392: | Line 364: | ||
| − | + | ||
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--标题--> | ||
<h2> | <h2> | ||
<span><span style="color:#7f1015">Abstract</span></span><hr /> | <span><span style="color:#7f1015">Abstract</span></span><hr /> | ||
| Line 402: | Line 386: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">This | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This | ||
| − | model | + | model is created to evaluate the effectiveness of initial design, and offers |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | guidelines on how the system can (or must) be improved. (You can go to <a href="/Team:NUDT_CHINA/Design">PROJECT. page</a> to see more.) The core idea is to simulate the process of producing the signal which can be detected, and draw a conclusion by obtaining the relationship between the signal intensity and the concentration of miRNA. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 420: | Line 397: | ||
<span><span style="color:#7f1015">Introduction </span></span><hr /> | <span><span style="color:#7f1015">Introduction </span></span><hr /> | ||
</h2> | </h2> | ||
| − | |||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| − | RCA model and a | + | create mathematical models of two aspects of our project, a RCA model and a |
| − | </span> | + | signal detection model.</span> |
</p> | </p> | ||
</br> | </br> | ||
| − | |||
| Line 439: | Line 414: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA | ||
| − | model | + | model is based on the Michaelis-Menten equation. We can obtain the relationship |
| − | concentration of miRNA and the number of stem-loop structures | + | between the concentration of miRNA and the number of stem-loop structures |
| − | through theoretical calculation, and our experimental results | + | through theoretical calculation, and we use our experimental results to |
calculate the parameters we introduced previously.</span> | calculate the parameters we introduced previously.</span> | ||
| Line 452: | Line 427: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In the |
| − | signal detection model | + | signal detection model, we combine a probability model with the kinetic equation |
| − | + | of enzymatic reaction, so we can obtain the relationship between the number of | |
stem-loop structures and the signal intensity under the premise of adding a | stem-loop structures and the signal intensity under the premise of adding a | ||
| − | certain amount of fusion proteins of dCas9 and split-HRP fragments | + | certain amount of the fusion proteins of dCas9 and split-HRP fragments. </span> |
| − | + | ||
| − | </span> | + | |
</p> | </p> | ||
</br> | </br> | ||
| Line 471: | Line 444: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">By | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By | ||
| − | integrating the two models, the impacts of the | + | integrating the two models, we can theoretically predict the impacts of the |
| − | the signal to noise ratio | + | molecule number of proteins on the signal to noise ratio and explain the trend |
| − | signal intensity | + | of the signal intensity variation against the change of the concentration of miRNA in our |
| − | work | + | wet-lab work. |
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 491: | Line 465: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> About model </span></span | + | <span><span style="color:#7f1015"> About model </span></span> |
</h3> | </h3> | ||
| Line 497: | Line 471: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1. MiRNA is not degraded throughout the reaction process.</span> |
| − | + | ||
| − | + | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 506: | Line 478: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">2. The two fusion proteins of dCas9 and split-HRP fragments have the |
| − | + | ||
| − | + | ||
same ability to combine with the stem-loop structure, and only when two | same ability to combine with the stem-loop structure, and only when two | ||
| Line 521: | Line 491: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">3. The number of stem-loop structures in each RCA product is equal |
| − | + | ||
| − | + | ||
under a certain reaction time. </span> | under a certain reaction time. </span> | ||
| Line 532: | Line 500: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">4. The enzymatic activity remains unchanged with time under the premise |
| − | + | ||
| − | + | of excessive amount of enzymes or a short-time reaction.</span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 543: | Line 510: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> About the data </span></span | + | <span><span style="color:#7f1015"> About the data </span></span> |
| − | + | ||
| − | + | ||
</h3> | </h3> | ||
| Line 551: | Line 516: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1. The data we obtain from wet-lab experiment are reliable.</span> |
| − | + | ||
| − | reliable.</span> | + | |
</p> | </p> | ||
</br> | </br> | ||
| Line 560: | Line 523: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">2. All the results are trustworthy in the process of statistical |
| − | + | processing and data calculation.</span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 574: | Line 537: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015">Notations </span></span | + | <span><span style="color:#7f1015">Notations </span></span> |
</h3> | </h3> | ||
| Line 586: | Line 549: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>Symbol </span> | + | <span style="font-family:"">Symbol </span> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>Definition </span> | + | <span style="font-family:"">Definition </span> |
</p> | </p> | ||
</td> | </td> | ||
| Line 598: | Line 561: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span> | + | <i><span style="font-family:"">x</span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK7"></a><a name="OLE_LINK6"></a><span>The concentration of</span><span> miRNA (pM)</span> | + | <a name="OLE_LINK7"></a><a name="OLE_LINK6"></a><span style="font-family:"">The |
| + | concentration of</span><span style="font-family:""> miRNA (pM)</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 610: | Line 574: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>C<sub>1</sub></span></i> | + | <i><span style="font-family:"">C<sub>1</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The concentration of initiated probe (Abbreviated | + | <span style="font-family:"">The |
| − | + | concentration of initiated probe (Abbreviated to iprobe)</span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 623: | Line 587: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>k<sub>1</sub></span></i> | + | <i><span style="font-family:"">k<sub>1</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK8"></a><span>A constant representing | + | <a name="OLE_LINK8"></a><span style="font-family:"">A constant representing the scale factor</span><span style="font-family:""></span> |
| − | + | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 636: | Line 599: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>K<sub>m </sub></span></i> | + | <i><span style="font-family:"">K<sub>m </sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>One of the characteristic constants of | + | <span style="font-family:"">One |
| − | + | of the characteristic constants of phi29 DNA polymerase</span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 649: | Line 612: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>V<sub>max</sub></span></i> | + | <i><span style="font-family:"">V<sub>max</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>One of the characteristic constants of phi29 | + | <span style="font-family:"">One |
| − | + | of the characteristic constants of phi29 DNA polymerase</span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 662: | Line 625: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>k<sub>2</sub></span></i> | + | <i><span style="font-family:"">k<sub>2</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span style="font-family:"">A |
| + | constant representing the scale factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 674: | Line 638: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>V</span></i> | + | <i><span style="font-family:"">V</span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The initial speed of RCA </span> | + | <span style="font-family:"">The initial |
| + | speed of RCA </span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 686: | Line 651: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK9"></a><i><span>n<sub>1</sub></span></i><i><sub><span></span></sub></i> | + | <a name="OLE_LINK9"></a><i><span style="font-family:"">n<sub>1</sub></span></i><i><sub><span style="font-family:""></span></sub></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The moles of RCA product</span> | + | <span style="font-family:"">The |
| + | moles of RCA product</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 698: | Line 664: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>n<sub>2</sub></span></i> | + | <i><span style="font-family:"">n<sub>2</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK10"></a><span>The number of | + | <a name="OLE_LINK10"></a><span style="font-family:"">The number of stem-loop structures in each RCA product</span><span style="font-family:""></span> |
| − | + | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 711: | Line 676: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>n</span></i> | + | <i><span style="font-family:"">n</span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The total amount of <a name="OLE_LINK23"></a><a name="OLE_LINK22"></a>stem-loop structures</span> | + | <span style="font-family:"">The total |
| + | amount of <a name="OLE_LINK23"></a><a name="OLE_LINK22"></a>stem-loop structures</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 723: | Line 689: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>N</span></i> | + | <i><span style="font-family:"">N</span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK41"></a><a name="OLE_LINK16"></a><span>The molecule number of fusion | + | <a name="OLE_LINK41"></a><a name="OLE_LINK16"></a><span style="font-family:"">The |
| − | + | molecule number of the fusion protein of dCas9 and split-HRP fragments</span><span style="font-family:""></span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 736: | Line 702: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>k<sub>3</sub></span></i> | + | <i><span style="font-family:"">k<sub>3</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span style="font-family:"">A |
| + | constant representing the scale factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 748: | Line 715: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>y<sub>1</sub></span></i> | + | <i><span style="font-family:"">y<sub>1</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The fluorescence | + | <span style="font-family:"">The fluorescence |
| − | DNA-dye-complex (RFU)</span> | + | intensity of DNA-dye-complex (RFU)</span> |
</p> | </p> | ||
</td> | </td> | ||
| Line 761: | Line 728: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>N<sub>1</sub></span></i> | + | <i><span style="font-family:"">N<sub>1</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The molecule number of fusion protein of | + | <span style="font-family:"">The |
| − | + | molecule number of the fusion protein of dCas9 and split-HRP fragments in the | |
| + | solution</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 774: | Line 742: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>N<sub>2</sub></span></i> | + | <i><span style="font-family:"">N<sub>2</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK45"></a><span>The molecule number | + | <a name="OLE_LINK45"></a><span style="font-family:"">The molecule number of the fusion protein of dCas9 and |
| − | + | split-HRP fragments binding with stem-loop structure</span><span style="font-family:""></span> | |
| − | + | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 788: | Line 755: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>k<sub>4</sub></span></i> | + | <i><span style="font-family:"">k<sub>4</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span style="font-family:"">A |
| + | constant representing the scale factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 800: | Line 768: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span>k<sub>5</sub></span></i> | + | <i><span style="font-family:"">k<sub>5</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span style="font-family:"">A |
| + | constant representing the scale factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 812: | Line 781: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:"">ρ</span></i><i><span style="font-family:""></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>Signal to noise ratio(Abbreviated to | + | <span style="font-family:"">Signal |
| − | + | to noise ratio(Abbreviated to SNR)</span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 825: | Line 794: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:"">I</span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The molecule number of formed intact HRP | + | <span style="font-family:"">The |
| − | + | molecule number of formed intact HRP proteins </span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 838: | Line 807: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:"">I<sub>1</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The molecule number of formed intact HRP | + | <span style="font-family:"">The |
| − | + | molecule number of formed intact HRP proteins in the solution</span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 851: | Line 820: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:"">I<sub>2</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK43"></a><span>The molecule number | + | <a name="OLE_LINK43"></a><span style="font-family:"">The molecule number of formed intact HRP proteins through |
| − | + | binding with stem-loop structure</span><span style="font-family:""></span> | |
</p> | </p> | ||
</td> | </td> | ||
| Line 864: | Line 833: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:"">t</span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>Reaction time</span> | + | <span style="font-family:"">Reaction |
| + | time</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 876: | Line 846: | ||
<td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:"">y<sub>2</sub></span></i> |
</p> | </p> | ||
</td> | </td> | ||
<td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The signal intensity (OD<sub>450</sub>)</span> | + | <span style="font-family:"">The |
| + | signal intensity (OD<sub>450</sub>)</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 888: | Line 859: | ||
</table> | </table> | ||
</p> | </p> | ||
| − | |||
| Line 894: | Line 864: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The RCA model </span></span | + | <span><span style="color:#7f1015"> The RCA model </span></span> |
| − | + | ||
| − | + | ||
</h3> | </h3> | ||
| Line 903: | Line 871: | ||
<!--正文--> | <!--正文--> | ||
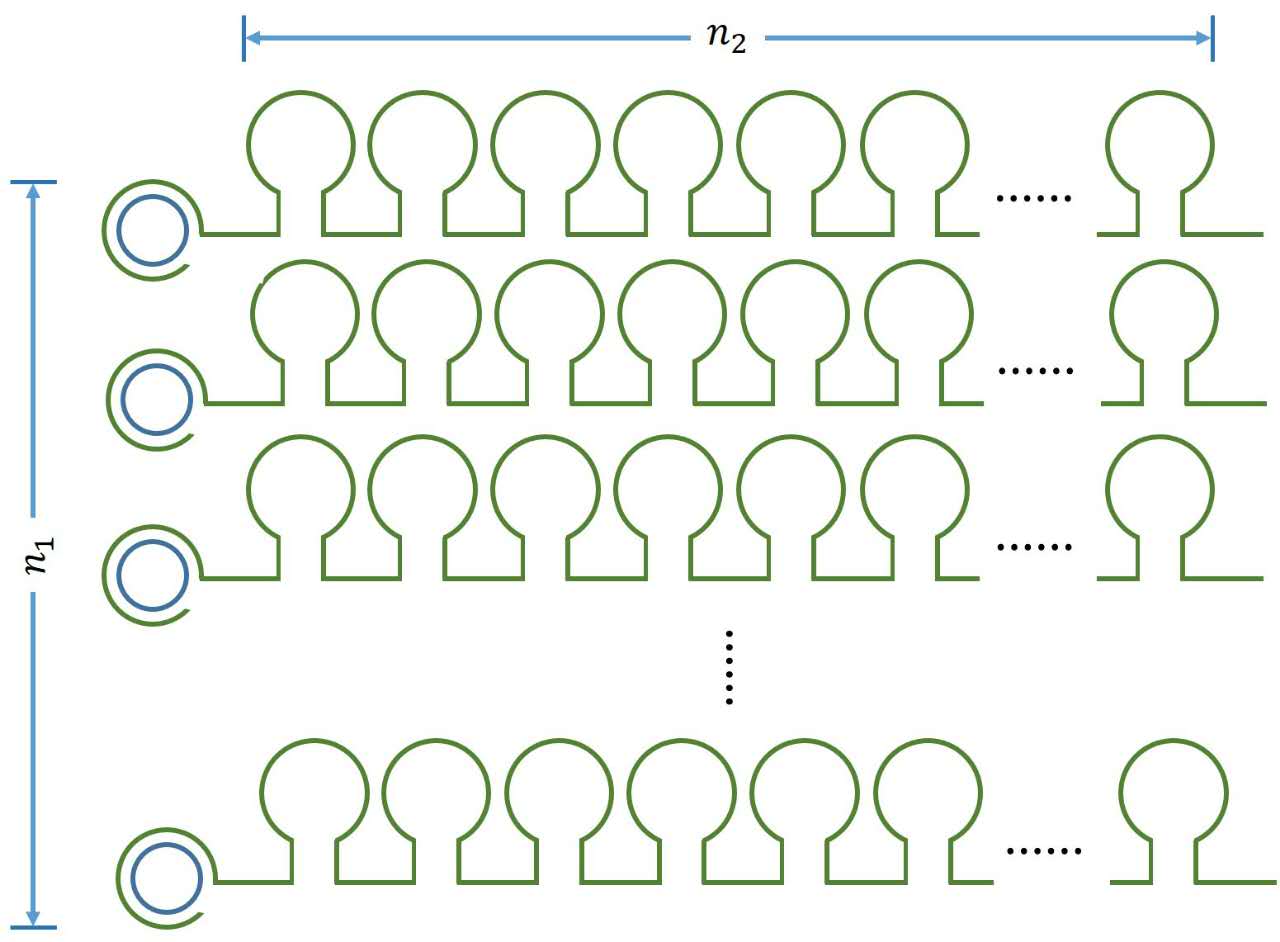
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, we | |
| + | |||
| + | assume that there is a linear relationship between the | ||
concentration of the initiated probes and miRNA as the combination reaction | concentration of the initiated probes and miRNA as the combination reaction | ||
| Line 909: | Line 879: | ||
between miRNA and probe occurs spontaneously<sup>1 </sup> . This can be written | between miRNA and probe occurs spontaneously<sup>1 </sup> . This can be written | ||
| − | as: | + | as:</span> |
| − | + | ||
| − | </span> | + | |
</p> | </p> | ||
</br> | </br> | ||
| + | |||
<!--插入图片--> | <!--插入图片--> | ||
| Line 958: | Line 927: | ||
obvious that there is a linear relationship between the molecule numbers of | obvious that there is a linear relationship between the molecule numbers of | ||
| − | iprobe-dN (n<sub>1 </sub>, reflects the molecule numbers of RCA product) and the initial | + | iprobe-dN (n<sub>1 </sub> |
| + | , reflects the molecule numbers of RCA product) and the initial speed of RCA | ||
| − | + | reaction. Notably, under the premise of excessive amount of enzymes, the | |
| − | + | extending of each RCA production is related to the reaction time, not the | |
| − | + | concentration of the iprobe. It is a linear relationship when we assume that the | |
| − | + | enzymatic activity remains unchanged with time, thus, the number of stem-loop | |
| − | + | structures (n<sub>2 </sub> | |
| + | ) in each RCA product is stable under the premise of a certain reaction time. | ||
| − | + | </span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 982: | Line 953: | ||
</br> | </br> | ||
| − | + | <!--正文--> | |
| − | + | ||
| − | <!-- | + | |
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | size:18px;">Figure 1. Schematic diagram</ | + | size:18px;"><b>Figure 1. Schematic diagram</b></span> |
</p> | </p> | ||
| − | |||
</br> | </br> | ||
| + | |||
| Line 1,025: | Line 994: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">And | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And | ||
| − | then, SYBR Green binds to DNA. The resulting DNA-dye-complex absorbs blue light | + | then, SYBR Green binds to DNA. The |
| + | resulting DNA-dye-complex absorbs blue light (<i>λ<sub>max</sub></i> = 497nm) | ||
| − | + | and emits green light (<i>λ<sub>max</sub></i> = 520nm). Thus, there is | |
| + | a linear relationship between the total amount of stem-loop structures and the | ||
| − | + | fluorescence | |
| − | + | intensity of DNA-dye-complex. This can be written as:</span> | |
| − | + | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,057: | Line 1,027: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model6.jpg| | + | [[File:T--NUDT_CHINA--model6.jpg|210px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,067: | Line 1,037: | ||
<span><span style="color:#7f1015"> The signal detection model | <span><span style="color:#7f1015"> The signal detection model | ||
| − | </span></span | + | </span></span> |
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, it is | |
| − | + | obvious that there is a linear relationship between | |
the molecule number of formed intact HRP proteins in the solution (consider as | the molecule number of formed intact HRP proteins in the solution (consider as | ||
| − | NOISE) and the molecule number of fusion | + | NOISE) and the molecule number of the fusion proteins in the solution. This can |
| − | written as:</span> | + | be written as:</span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,099: | Line 1,069: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, | ||
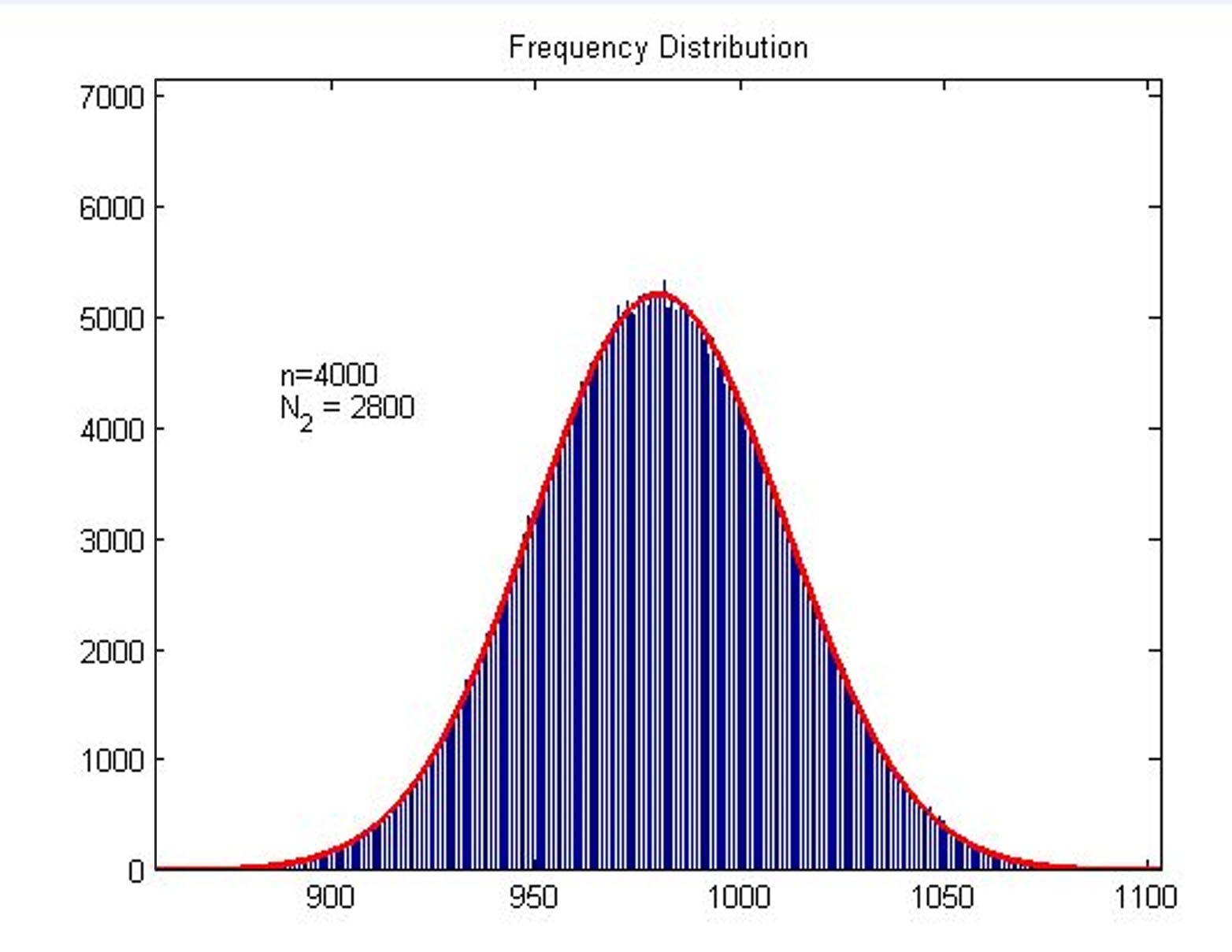
| − | we | + | we build a probability model to find the expression of the molecule number of |
formed intact HRP proteins (fused with dCas9) through binding with stem-loop | formed intact HRP proteins (fused with dCas9) through binding with stem-loop | ||
| Line 1,107: | Line 1,077: | ||
structures and the molecule number of DNA-bound fusion proteins. We obtain the | structures and the molecule number of DNA-bound fusion proteins. We obtain the | ||
| − | result by Monte Carlo method. </span> | + | result by Monte Carlo method. </span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,122: | Line 1,092: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | size:18px;">Figure2. Frequency Distribution | + | size:18px;"><b>Figure2. Frequency Distribution</b> Plot of frequency |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | distribution of the molecule number of formed intact HRP proteins through | |
| + | |||
| + | binding with stem-loop structure. </span> | ||
| − | |||
| − | |||
</br> | </br> | ||
| Line 1,143: | Line 1,110: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | size:18px;">Figure 3. The result of the probability model | + | size:18px;"><b>Figure 3. The result of the probability model</b> Plot of the |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | expect value of the molecule number of formed intact proteins against the total | |
| − | + | amount of stem-loop structures, carried out at 0.1, 0.3, 0.5, 0.7, 0.9 of | |
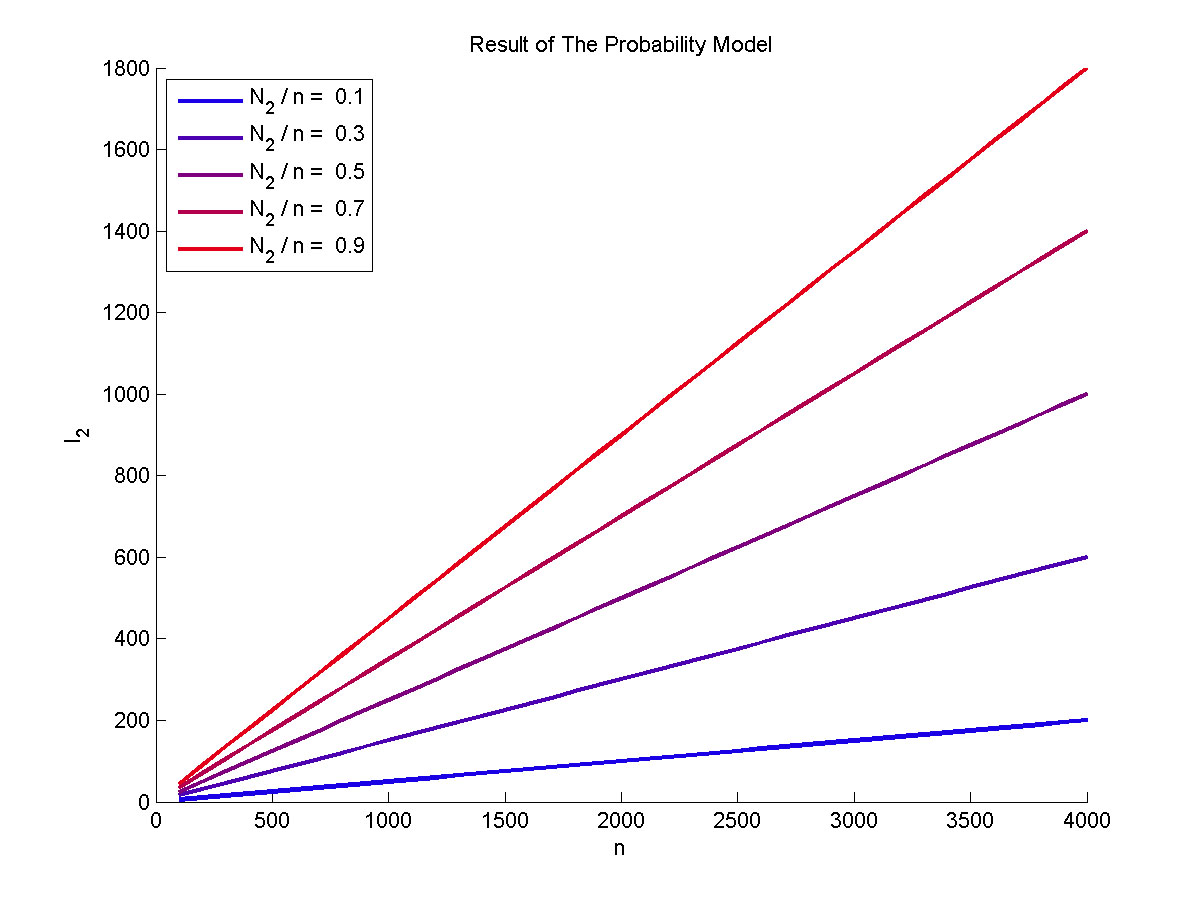
| − | + | N<sub>2</sub> to n ratio.</span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,171: | Line 1,135: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model8.jpg| | + | [[File:T--NUDT_CHINA--model8.jpg|70px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,179: | Line 1,143: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">As for | <span style="line-height:2;font-family:Perpetua;font-size:18px;">As for | ||
| − | + | <i>N<sub>2</sub></i>, | |
| − | + | which refers to the molecule number of the fusion proteins binding with | |
| − | loop structure. The equation is formulated based on the limiting case. The | + | stem-loop structure. The equation is formulated based on the limiting case. The |
| − | + | ||
equation can be written as:</span> | equation can be written as:</span> | ||
</p> | </p> | ||
| Line 1,192: | Line 1,155: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model9.jpg| | + | [[File:T--NUDT_CHINA--model9.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,201: | Line 1,164: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">And | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And | ||
| − | then, we can obtain the expressions for ρ and I separately.</span> | + | then, we can obtain the expressions for <i>ρ</i></span> <span>and<i> I</i> |
| + | |||
| + | separately.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,210: | Line 1,175: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model10.jpg| | + | [[File:T--NUDT_CHINA--model10.jpg|50px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,223: | Line 1,188: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Notably, under | |
| − | + | the premise of excessive substrates, | |
| + | it is a linear relationship between the reaction rate and the concentration of | ||
| + | enzymes when we assume that the enzymatic activity remain unchanged with time, | ||
| + | thus, we can obtain the expression of the signal intensity at <i>t<sub>th</sub> | ||
| − | + | </i>time-step. This is | |
| − | + | written as:</span> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | as:</span> | + | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,242: | Line 1,205: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model12.jpg| | + | [[File:T--NUDT_CHINA--model12.jpg|70px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,252: | Line 1,215: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">In | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In | ||
| − | summary, the relationship between the number of stem-loop structures and | + | summary, the relationship between the number of stem-loop structures and SNR |
| − | + | under the premise of adding a certain amount of the fusion proteins can be | |
| − | + | written as:</span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,277: | Line 1,240: | ||
relationship between the number of Stem-loop structures and the signal intensity | relationship between the number of Stem-loop structures and the signal intensity | ||
| − | under the premise of adding a certain amount of fusion proteins | + | under the premise of adding a certain amount of the fusion proteins can be |
| − | as:</span> | + | written as:</span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,288: | Line 1,251: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model14.jpg| | + | [[File:T--NUDT_CHINA--model14.jpg|280px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,296: | Line 1,259: | ||
<span><span style="color:#7f1015"> The calculation of the constants | <span><span style="color:#7f1015"> The calculation of the constants | ||
| − | </span></span | + | </span></span> |
</h3> | </h3> | ||
| Line 1,303: | Line 1,266: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">To | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To | ||
| − | simplify the equation (1), the constants | + | simplify the equation (1), the constants |
| − | + | can be integrated. As can be seen from the above table, <i>k<sub>1</sub></i>, | |
| − | from the above table, | + | |
| − | + | <i>k<sub>2</sub></i>, <i>k<sub>3</sub></i><sub> </sub>are | |
| + | constants representing the scale factor, <i>K<sub>m</sub></i> and | ||
| − | + | <i>V<sub>max</sub></i> are | |
| + | characteristic constants of phi 29 DNA polymerase, and <i>n<sub>2</sub></i>, | ||
| − | under the premise of a certain reaction time. Thus, define two constants, mark | + | which refers to the number of stem-loop structures |
| + | in each RCA product, is stable under the premise of a certain reaction time. | ||
| + | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the equation | ||
| − | + | can be | |
| + | simplified as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,320: | Line 1,287: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model15.jpg| | + | [[File:T--NUDT_CHINA--model15.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,326: | Line 1,293: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
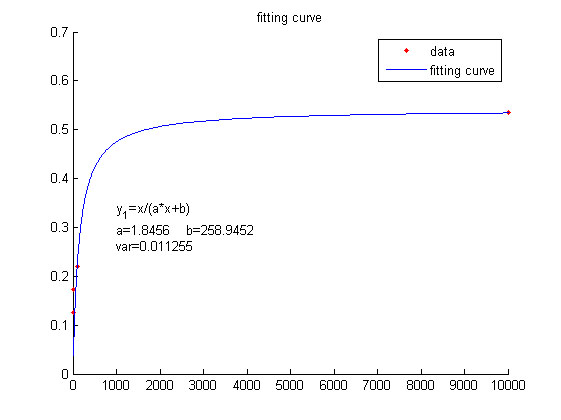
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We use |
| − | this equation to fit the data points obtained through experiments. (Figure | + | this equation to fit the data points obtained through experiments. (Figure 2. |
| − | the | + | (D)in the <a href="/Team:NUDT_CHINA/Results">RESULTS.page</a>) The fitting |
| + | |||
| + | curve is shown below.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,342: | Line 1,311: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | size:18px;">Figure 4. Fitting curve | + | size:18px;"><b>Figure 4. Fitting curve</b> Fitting curve of fluorescence |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | intensity of DNA-dye-complex (RFU) against the concentrations of miRNA | |
| − | + | (pM).</span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,363: | Line 1,329: | ||
follows: | follows: | ||
| − | + | <i>k<sub>3</sub> </i>=8.35*10<sup>-16 </sup> | |
| + | |||
| + | (<i>n</i>=<i>y<sub>1</sub></i>/<i>k<sub>3</sub></i>); <i>k<sub>4</sub></i> | ||
| + | |||
| + | =0.038;<i> k<sub>5</sub>t</i>=5.9*10<sup>-12</sup>. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 1,373: | Line 1,343: | ||
<span><span style="color:#7f1015"> Results and Analysis | <span><span style="color:#7f1015"> Results and Analysis | ||
| − | </span></span | + | </span></span> |
</h3> | </h3> | ||
| Line 1,380: | Line 1,350: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">By | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By | ||
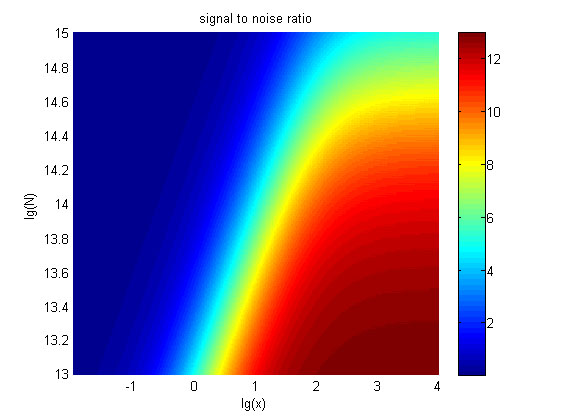
| − | integrating the two models, we can obtain the relationships between the | + | integrating the two models, we can obtain the relationships between SNR, the |
| − | + | signal intensity respectively and the concentration of miRNA under different | |
| − | + | addition amount of fusion proteins. | |
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,403: | Line 1,374: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | dimensional map of signal | + | size:18px;"><b>Figure |
| + | 5. The signal intensity (OD<sub>450</sub>)</b> Three-dimensional map of signal | ||
| − | additional amount of fusion proteins. </span> | + | intensity (OD<sub>450</sub>) |
| + | against miRNA concentration(pM) and additional amount of fusion proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,422: | Line 1,390: | ||
exists a monotonous relation between the concentration of miRNA and the signal | exists a monotonous relation between the concentration of miRNA and the signal | ||
| − | intensity when the value of the molecule number of fusion proteins is | + | intensity when the value of the molecule number of the fusion proteins is |
| − | large. While the relationship does not hold when the value of the | + | relatively large. While the relationship does not hold when the value of the |
| − | number of fusion proteins is relatively small. And the signal | + | molecule number of the fusion proteins is relatively small. And the signal |
| − | increases as the value of the molecule number of fusion proteins | + | intensity increases as the value of the molecule number of the fusion proteins |
increases.</span> | increases.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig6.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;"><b>Figure 6. The result of SNR</b> Three-dimensional map of signal | ||
| + | |||
| + | to noise ratio against miRNA concentration (pM) and additional amount of fusion | ||
| + | |||
| + | proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,439: | Line 1,426: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">The | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| − | + | signal-to-noise ratio decreases as the value of the molecule number of fusion | |
| − | + | proteins increases. </span> | |
| + | </p> | ||
| + | </br> | ||
| − | + | ||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> Test of model | ||
| + | |||
| + | </span></span> | ||
| + | </h3> | ||
| + | |||
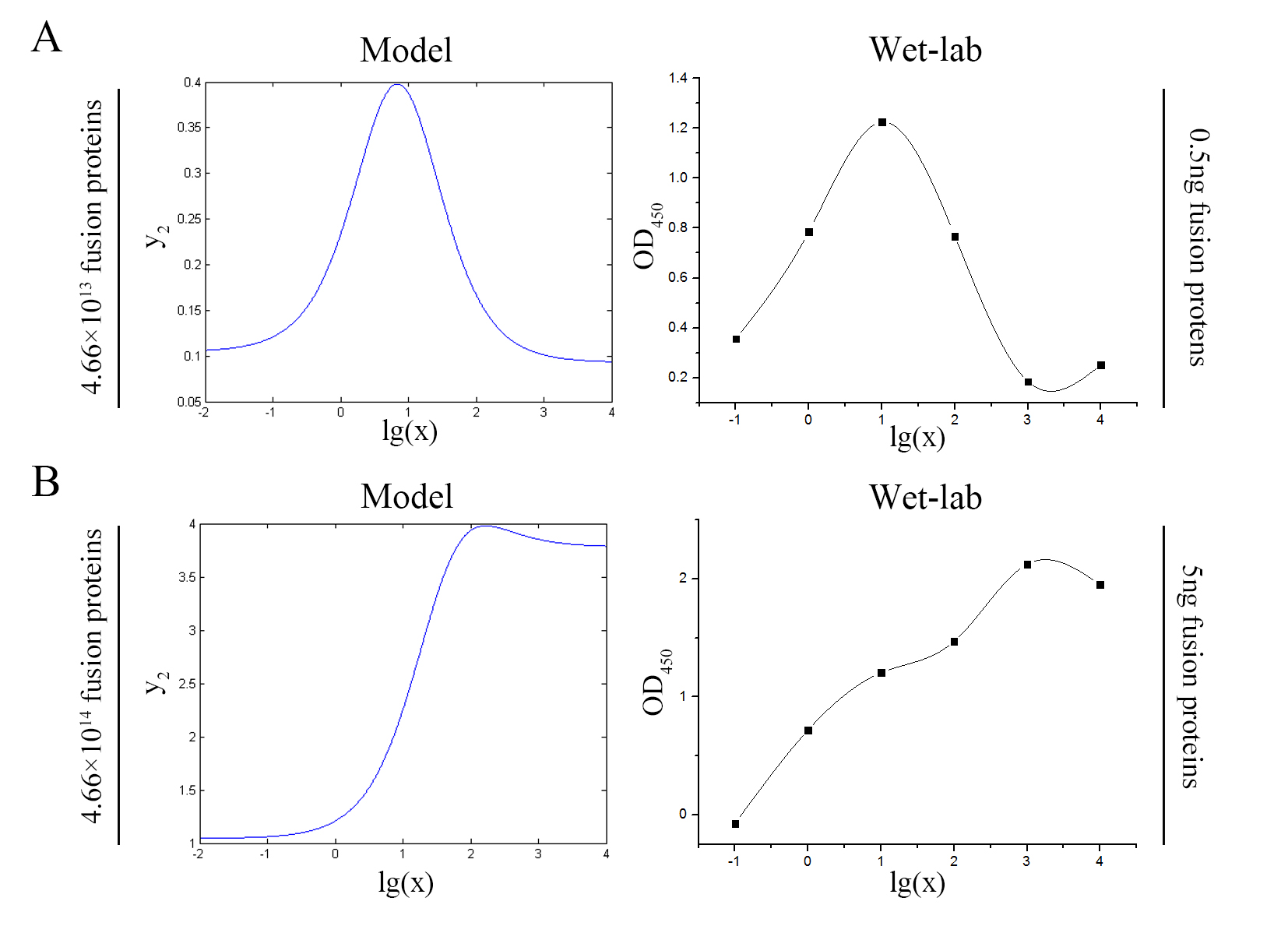
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Our | ||
| + | |||
| + | model predicts the relationship between the signal intensity and the | ||
| + | |||
| + | concentration of miRNA under the premise of adding a certain amount of the | ||
| + | |||
| + | fusion proteins of dCas9 and split-HRP fragments. So we can test if our model is | ||
| + | |||
| + | reasonable by comparing the curves predicted by the model with those obtained | ||
| + | |||
| + | from the experiment. The results are shown below. | ||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| + | |||
| Line 1,451: | Line 1,462: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA-- | + | [[File:T--NUDT_CHINA--modelfig7-2.jpg|700px|center]] |
<html> | <html> | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | size:18px;">Figure | + | size:18px;"><b>Figure 7. Test of model</b> Plot of the signal intensity (OD<sub>450</sub>) against miRNA concentration (pM) predicted by the model and experimented by wet-lab. (A) The value of the molecule number of each fusion protein added to the system is set to be 4.66e+13. The mass of each fusion protein added to the system in wet-lab is 0.5 ng. (B) The value of the molecule number of each fusion protein added to the system is set to be 4.66e+14. The mass of each fusion protein added to the system in wet-lab is 5 ng.</span> |
</p> | </p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</br> | </br> | ||
| + | |||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We can learn | |
| − | + | from the figure above that the general trend of curves predicted by the model agree well with the experimental results. </span> | |
| − | + | ||
| − | + | ||
</p> | </p> | ||
</br> | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> Model extension | ||
| + | |||
| + | </span></span> | ||
| + | </h3> | ||
<!--正文--> | <!--正文--> | ||
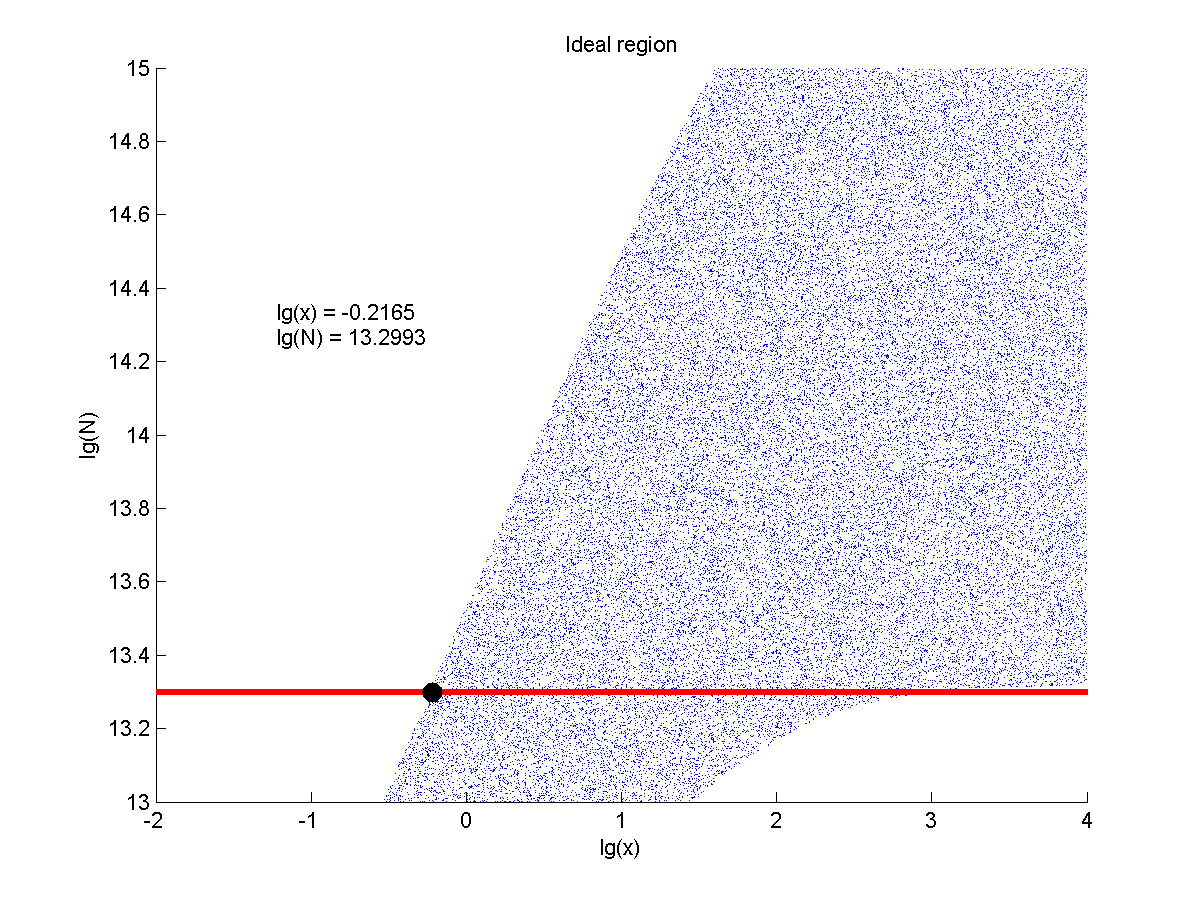
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Furthermore, we | |
| − | + | ||
| − | size:18px;">Furthermore, we | + | |
| − | when the | + | consider that it is conducive to signal detection when the value of signal |
| − | than two. We obtain the ideal region through | + | intensity and SNR are both greater than two. We obtain the ideal region through |
| − | below. </span> | + | calculation, which is shown as below. </span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,503: | Line 1,513: | ||
</br> | </br> | ||
<p> | <p> | ||
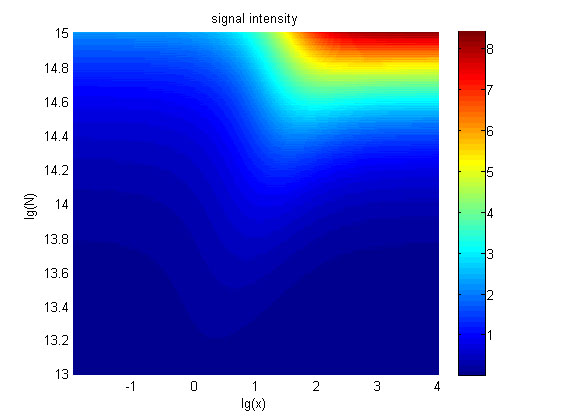
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | size:18px;">Figure | + | size:18px;"><b>Figure 8. Ideal region</b> Region of qualified logarithm of |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | concentrations of miRNA (pM) and logarithm of the additional amount of fusion | |
| − | + | proteins.</span> | |
</p> | </p> | ||
| + | |||
</br> | </br> | ||
| Line 1,522: | Line 1,530: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">When | <span style="line-height:2;font-family:Perpetua;font-size:18px;">When | ||
| − | the value of N is set to | + | the value of N is set to e+13.3, the range of the value of x contained in the |
| − | ideal region is | + | ideal region is a maximum, which means when the value of the molecule number of |
| − | + | the fusion proteins is e+13.3, the concentration range of miRNA that can be | |
| − | + | detected is maximum. </span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,549: | Line 1,557: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The general trend of curves predicted by the model are in good agreement with the experimental |
| − | + | ||
| − | + | ||
| − | + | results, thus indicating that our model is reasonable.</span> | |
| − | + | ||
| − | + | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,563: | Line 1,567: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">The | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| − | value of the molecule number of fusion protein of dCas9 and split-HRP fragments | + | value of the molecule number of the fusion protein of dCas9 and split-HRP |
| + | |||
| + | fragments not only affects the signal-to-noise ratio, but also the signal | ||
| − | + | intensity. So we need to weigh its impact on both to select the optimal | |
| − | + | solution.</span> | |
</p> | </p> | ||
</br> | </br> | ||
| Line 1,576: | Line 1,582: | ||
<span style="line-height:2;font-family:Perpetua;font-size:18px;">The | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| − | concentration range of miRNA | + | concentration range of miRNA that can be detected is a maximum when we set the |
| − | + | value of the molecule number of the fusion proteins to be e+13.3, building on | |
| − | + | which, our wet-lab protocol could be optimized in our future work.</span> | |
| − | + | ||
| − | + | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,593: | Line 1,597: | ||
<!--正文--> | <!--正文--> | ||
| − | <p | + | <p > |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1 Deng, R. <i>et al</i>.Toehold-initiated rolling circleamplification for visualizing individual microRNAs in situ in single cells. <i>Angew Chem Int Ed Engl </i>53, 2389-2393,doi:10.1002/anie.201309388 (2014).</span> | |
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 1,618: | Line 1,635: | ||
</div> | </div> | ||
<!-- end #content-wrap --> | <!-- end #content-wrap --> | ||
| − | + | <div style="border:solid 1px rgb(10,31,84);background-color:rgb(10,31,84);"> | |
| − | + | <div class="row footer-link" style="border:solid 0px red;margin-top:100px;"> | |
| − | + | <style>.menu-link-container ul li{ list-style:none; } .menu-link-container a{ color:white; font-size:14px;line-height:26px; } | |
| − | + | .menu-link-container a:hover{ | |
| − | + | text-decoration:underline; | |
| − | + | } #c_menu9909 .medium-6{ text-align:center; width:auto; border:solid 0px red; } #c_menu9909 .medium-6 h5{color:white;}#c_menu9909 .medium-6{height:315px;border:solid 0px red;}</style> | |
| − | + | <div id="c_menu9909"> | |
| − | + | <!--div class="medium-6 columns" style="width:auto;"> | |
| − | + | <h5 class="column-header">HOME</h5> | |
| − | + | <div class="menu-link-container"></div></div--> | |
| − | + | <div class="medium-6 columns" style="width:auto;"> | |
| − | + | <h5 class="column-header">TEAM</h5> | |
| − | + | <div class="menu-link-container"> | |
| − | + | ||
| − | + | ||
| − | + | <a href="/Team:NUDT_CHINA/Team">Team</a><br/> | |
| − | + | ||
| − | + | <a href="/Team:NUDT_CHINA/Collaborations">Collaborations</a><br/> | |
| − | + | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">PROJECT</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Description">Description</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Design">Design</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Experiments">Experiments</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Proof">Proof of Concept</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Demonstrate">Demonstrate</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Results">Results</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Notebook">Notebook</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">PARTS</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Parts">Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Basic_Part">Basic Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Composite_Part">Composite Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Part_Collection">Part Collection</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <a href="/Team:NUDT_CHINA/Safety" style="text-decoration:none;"> | ||
| + | <h5 class="column-header">SAFETY</h5></a> | ||
| + | |||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <a href="/Team:NUDT_CHINA/Attributions" style="text-decoration:none;"> | ||
| + | <h5 class="column-header">ATTRIBUTIONS</h5></a> | ||
| + | |||
| + | </div> | ||
| + | <!--div class="medium-6 columns" style="width:auto;"> | ||
| + | <A href="/Team:NUDT_CHINA/Attributions" style="text-decoration:none;"><h5 class="column-header">ATTRIBUTIONS</h5></A> | ||
| + | <div class="menu-link-container"></div></div--> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">HUMAN | ||
| + | <br>PRACTICES</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Human_Practices">Human Practices</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/HP/Silver">Silver</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/HP/Gold">Gold</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Integrated_Practices">Integrated Practices</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Engagement">Engagement</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">AWARDS</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Model">Model</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:0px;height:0px;"></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
<footer id="footer"> | <footer id="footer"> | ||
<div class="row footer-link"> | <div class="row footer-link"> | ||
| Line 1,768: | Line 1,786: | ||
--> | --> | ||
| − | + | <!-- to top edit by inksci---> | |
| + | <style> | ||
| + | #to_top{ | ||
| + | position:fixed; | ||
| + | bottom:100px; | ||
| + | right:0px; | ||
| + | width:60px; | ||
| + | height:60px; | ||
| + | font-size:14px; | ||
| + | line-height:60px; | ||
| + | text-align:center; | ||
| + | background-color:black; | ||
| + | color:rgb(246,168,0); | ||
| + | cursor:pointer; | ||
| + | } | ||
| + | </style> | ||
| + | <div onclick="top_it()" id="to_top"> | ||
| + | TOP | ||
| + | </div> | ||
| + | <script type="text/javascript"> | ||
| + | var oTop = document.getElementById("to_top"); | ||
| + | oTop.style.display="none"; | ||
| + | var top_break=false; | ||
| + | /** | ||
| + | * 回到页面顶部 | ||
| + | * @param acceleration 加速度 | ||
| + | * @param time 时间间隔 (毫秒) | ||
| + | **/ | ||
| + | var save_scrolltop=100000000000000; | ||
| + | window.onscroll = function(){ | ||
| + | |||
| + | |||
| + | var scrolltop = document.documentElement.scrollTop || document.body.scrollTop; | ||
| + | if(scrolltop>300){ | ||
| + | oTop.style.display="block"; | ||
| + | }else{ | ||
| + | oTop.style.display="none"; | ||
| + | } | ||
| + | if(scrolltop>save_scrolltop) | ||
| + | { | ||
| + | top_break=true; | ||
| + | } | ||
| + | save_scrolltop=scrolltop; | ||
| + | } | ||
| + | function top_it(){ | ||
| + | top_break=false; | ||
| + | goTop();return false; | ||
| + | } | ||
| + | function goTop(acceleration, time) { | ||
| + | if(top_break){ | ||
| + | return; | ||
| + | } | ||
| + | oTop.style.display="none"; | ||
| + | acceleration = acceleration || 0.1; | ||
| + | time = time || 16; | ||
| + | |||
| + | var x1 = 0; | ||
| + | var y1 = 0; | ||
| + | var x2 = 0; | ||
| + | var y2 = 0; | ||
| + | var x3 = 0; | ||
| + | var y3 = 0; | ||
| + | |||
| + | if (document.documentElement) { | ||
| + | x1 = document.documentElement.scrollLeft || 0; | ||
| + | y1 = document.documentElement.scrollTop || 0; | ||
| + | } | ||
| + | if (document.body) { | ||
| + | x2 = document.body.scrollLeft || 0; | ||
| + | y2 = document.body.scrollTop || 0; | ||
| + | } | ||
| + | var x3 = window.scrollX || 0; | ||
| + | var y3 = window.scrollY || 0; | ||
| + | |||
| + | // 滚动条到页面顶部的水平距离 | ||
| + | var x = Math.max(x1, Math.max(x2, x3)); | ||
| + | // 滚动条到页面顶部的垂直距离 | ||
| + | var y = Math.max(y1, Math.max(y2, y3)); | ||
| + | |||
| + | // 滚动距离 = 目前距离 / 速度, 因为距离原来越小, 速度是大于 1 的数, 所以滚动距离会越来越小 | ||
| + | var speed = 1 + acceleration; | ||
| + | window.scrollTo(Math.floor(x / speed), Math.floor(y / speed)); | ||
| + | |||
| + | // 如果距离不为零, 继续调用迭代本函数 | ||
| + | if(x > 0 || y > 0) { | ||
| + | var invokeFunction = "goTop(" + acceleration + ", " + time + ")"; | ||
| + | window.setTimeout(invokeFunction, time); | ||
| + | } | ||
| + | } | ||
| + | </script> | ||
| + | <!-- to top edit by inksci---> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 13:34, 19 October 2016
TOP