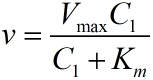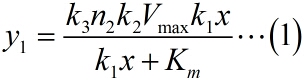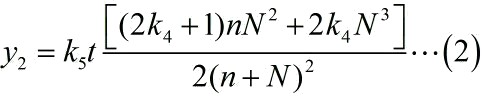Zhuchushu13 (Talk | contribs) |
Zhuchushu13 (Talk | contribs) |
||
| (36 intermediate revisions by 2 users not shown) | |||
| Line 69: | Line 69: | ||
<meta name="HandheldFriendly" content="True" /> | <meta name="HandheldFriendly" content="True" /> | ||
<meta name="MobileOptimized" content="320" /> | <meta name="MobileOptimized" content="320" /> | ||
| − | + | <meta name="viewport" content="width=device-width, initial-scale=1.0",minimum-scale=0.2, maximum-scale=2.0, user-scalable=yes" /> | |
<meta content="on" http-equiv="cleartype" /> | <meta content="on" http-equiv="cleartype" /> | ||
<meta property="og:title" content="Home" /> | <meta property="og:title" content="Home" /> | ||
| Line 88: | Line 88: | ||
<div class="inner-wrap"> | <div class="inner-wrap"> | ||
<!-- Off Canvas Menu --> | <!-- Off Canvas Menu --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- close the off-canvas menu --> | <!-- close the off-canvas menu --> | ||
<a class="exit-off-canvas"></a> | <a class="exit-off-canvas"></a> | ||
| Line 152: | Line 115: | ||
<a class="nav-global__link" title="" href="#">AWARDS</a></li> | <a class="nav-global__link" title="" href="#">AWARDS</a></li> | ||
</ul> | </ul> | ||
| − | <style>#menu06{width:200px;right:0px;} #menu07{width:200px;right:0px;} | + | <style> |
| + | .content{ | ||
| + | box-shadow: 6px 6px 3px #888888; | ||
| + | } | ||
| + | #menu06{width:200px;right:0px;} #menu07{width:200px;right:0px;} | ||
#menu01 .arrow-up{margin-right:6.429rem;} | #menu01 .arrow-up{margin-right:6.429rem;} | ||
#menu02 .arrow-up{margin-right:90px;} #menu03 .arrow-up{margin-right:90px;} #menu06 .arrow-up{margin-right:120px;} #menu07 .arrow-up{margin-right:27px;} #menu01{width:200px;right:31rem;} #menu02{width:200px;right:27rem;} #menu03{width:200px;right:21.3rem;} | #menu02 .arrow-up{margin-right:90px;} #menu03 .arrow-up{margin-right:90px;} #menu06 .arrow-up{margin-right:120px;} #menu07 .arrow-up{margin-right:27px;} #menu01{width:200px;right:31rem;} #menu02{width:200px;right:27rem;} #menu03{width:200px;right:21.3rem;} | ||
| Line 322: | Line 289: | ||
</div> | </div> | ||
</nav> | </nav> | ||
| − | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } }</style> | + | <style>.nav-global ul li a{ font-size:0.85rem; } @media screen and (max-width: 865px) { .nav-global ul li a{ font-size:0.85rem; } } |
| + | |||
| + | #feature{ | ||
| + | background-size:100%; | ||
| + | } | ||
| + | @media screen and (max-width: 1200px) { | ||
| + | #feature{ | ||
| + | background-size:1200px; | ||
| + | } | ||
| + | } | ||
| + | </style> | ||
<div class="breadcrumb-share-wrap"> | <div class="breadcrumb-share-wrap"> | ||
| Line 335: | Line 312: | ||
<!-- end .row --></div> | <!-- end .row --></div> | ||
<!-- end #header-outer-wrap--> | <!-- end #header-outer-wrap--> | ||
| − | <div id="feature" class="row collapse text-light feature-footer-active"> | + | <div id="feature" class="row collapse text-light feature-footer-active" style="background-color: rgb(3,13,38);background-image:url(https://static.igem.org/mediawiki/2016/thumb/3/3f/NUDT_CHINA2016_FILES-img-banner-3.0.png/800px-NUDT_CHINA2016_FILES-img-banner-3.0.png);background-position:0px -350px;background-repeat:no-repeat;"> |
<div class="inner-wrap medium-24 columns"> | <div class="inner-wrap medium-24 columns"> | ||
<!--TYPO3SEARCH_begin--> | <!--TYPO3SEARCH_begin--> | ||
| Line 341: | Line 318: | ||
<div class="carousel " id="slideshow-4811" data-cycle-log="false" data-cycle-auto-height="calc" data-cycle-prev="#prev-4811" data-cycle-next="#next-4811" data-cycle-pager="#cycle-pager-4811" data-cycle-easing="easeInOutQuad" data-cycle-fx="scrollVertUp" data-cycle-center-horz="true" data-cycle-speed="1000" data-cycle-timeout="8000" data-cycle-paused="true" data-cycle-slides="> div.slide"> | <div class="carousel " id="slideshow-4811" data-cycle-log="false" data-cycle-auto-height="calc" data-cycle-prev="#prev-4811" data-cycle-next="#next-4811" data-cycle-pager="#cycle-pager-4811" data-cycle-easing="easeInOutQuad" data-cycle-fx="scrollVertUp" data-cycle-center-horz="true" data-cycle-speed="1000" data-cycle-timeout="8000" data-cycle-paused="true" data-cycle-slides="> div.slide"> | ||
<div class="slide first"> | <div class="slide first"> | ||
| − | <link rel="stylesheet" href="/Team | + | <link rel="stylesheet" href="/Team--NUDT_CHINA/CSS3.css" /> |
<!-- Quick initialize with theme color from first slide --> | <!-- Quick initialize with theme color from first slide --> | ||
<div class="row collapse " id="c6601" data-theme-class="bh-color__theme-c6601"> | <div class="row collapse " id="c6601" data-theme-class="bh-color__theme-c6601"> | ||
<div class="columns"> | <div class="columns"> | ||
| − | <section class="big-header bh__height-size-3 text-light bh__video"> | + | <section class="big-header bh__height-size-3 text-light bh__video" style="height:300px;"> |
| − | <div class="bh__background bh__background-color | + | <div class="bh__background bh__background-color "></div> |
| − | <div class="bh__content-wrap"> | + | <div class="bh__content-wrap" style="height:120px;"> |
<div class="bh__content "> | <div class="bh__content "> | ||
<div class="bh__content-inner-wrap"> | <div class="bh__content-inner-wrap"> | ||
<!--h3 class="bh__subtitle"><a href="http#/explore/whats-here/exhibits/brick-by-brick/"><span class="line-wrap"><span class="line"><span class="bh__info">Exhibit / </span>Brick by Brick</span></span></a></h3--> | <!--h3 class="bh__subtitle"><a href="http#/explore/whats-here/exhibits/brick-by-brick/"><span class="line-wrap"><span class="line"><span class="bh__info">Exhibit / </span>Brick by Brick</span></span></a></h3--> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<h2 style="font-size:36px;" class="bh__title "> | <h2 style="font-size:36px;" class="bh__title "> | ||
<!--<a href="#">--> | <!--<a href="#">--> | ||
| − | + | <span style="line-height:46px;" class="line-wrap"> | |
| − | + | <span style="line-height:46px;" class="line">Development of A Novel</span></span> | |
| − | + | <span style="line-height:46px;" class="line-wrap"> | |
| − | + | <span style="line-height:46px;" class="line">Blood-MicroRNA Handy Detection System with CRISPR</span></span> | |
| − | <!--</a>--> | + | <!--</a>--></h2> |
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 378: | Line 345: | ||
<div class="cycle-pager" id="cycle-pager-4811"></div> | <div class="cycle-pager" id="cycle-pager-4811"></div> | ||
</div> | </div> | ||
| − | + | </div> | |
| − | + | </div> | |
<!-- end #feature --> | <!-- end #feature --> | ||
<!-- end .columns --></div> | <!-- end .columns --></div> | ||
| − | <div style="margin-top: | + | <div style="margin-top:20px;" id="content-wrap-ink"><!-- adjust in add_JS --> |
| − | <div class="row footer-link" style=""> | + | <div class="row footer-link" style=""> |
| + | <div style="text-align:right;"><h5> | ||
| + | <a style="color:rgb(10,31,84);" href="/Team:NUDT_CHINA">HomePage</a> • | ||
| + | <!-- 修改这里!! -->AWARDS • | ||
| + | <a style="color:rgb(10,31,84);" href="/Team:NUDT_CHINA/Model"><!-- 修改这里!! -->Model</a> | ||
| + | </h5><hr style="width:40%;margin-left:60%;border-top:1px solid rgb(10,31,84);" /></div> | ||
| Line 392: | Line 364: | ||
| − | + | ||
| − | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--标题--> | ||
<h2> | <h2> | ||
<span><span style="color:#7f1015">Abstract</span></span><hr /> | <span><span style="color:#7f1015">Abstract</span></span><hr /> | ||
| Line 400: | Line 384: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This model is created to evaluate the effectiveness of initial design, and offers guidelines | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This |
| + | |||
| + | model is created to evaluate the effectiveness of initial design, and offers | ||
| + | |||
| + | guidelines on how the system can (or must) be improved. (You can go to <a href="/Team:NUDT_CHINA/Design">PROJECT. page</a> to see more.) The core idea is to simulate the process of producing the signal which can be detected, and draw a conclusion by obtaining the relationship between the signal intensity and the concentration of miRNA. | ||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 412: | Line 401: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We create mathematical models of two aspects of our project, a RCA model and a signal detection model.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | create mathematical models of two aspects of our project, a RCA model and a | ||
| + | |||
| + | signal detection model.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 419: | Line 412: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA model is based on the Michaelis-Menten equation. We can obtain the relationship between the concentration of miRNA and the number of stem-loop structures through theoretical calculation, and we use our experimental results to calculate the parameters we introduced previously.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA |
| + | |||
| + | model is based on the Michaelis-Menten equation. We can obtain the relationship | ||
| + | |||
| + | between the concentration of miRNA and the number of stem-loop structures | ||
| + | |||
| + | through theoretical calculation, and we use our experimental results to | ||
| + | |||
| + | calculate the parameters we introduced previously.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 426: | Line 427: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In the signal detection model, we combine a probability model with the kinetic equation of enzymatic reaction, so we can obtain the relationship between the number of stem-loop structures and the signal intensity under the premise of adding a certain amount of the fusion proteins of dCas9 and split-HRP fragments. </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In the |
| + | |||
| + | signal detection model, we combine a probability model with the kinetic equation | ||
| + | |||
| + | of enzymatic reaction, so we can obtain the relationship between the number of | ||
| + | |||
| + | stem-loop structures and the signal intensity under the premise of adding a | ||
| + | |||
| + | certain amount of the fusion proteins of dCas9 and split-HRP fragments. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 433: | Line 442: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By integrating the two models, we can theoretically predict the impacts of the molecule number of proteins on the signal to noise ratio and | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By |
| + | |||
| + | integrating the two models, we can theoretically predict the impacts of the | ||
| + | |||
| + | molecule number of proteins on the signal to noise ratio and explain the trend | ||
| + | |||
| + | of the signal intensity variation against the change of the concentration of miRNA in our | ||
| + | |||
| + | wet-lab work. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 441: | Line 458: | ||
<!--标题--> | <!--标题--> | ||
<h2> | <h2> | ||
| − | <span><span style="color:#7f1015"> Assumption and Justification </span></span><hr /> | + | <span><span style="color:#7f1015"> Assumption and Justification |
| + | |||
| + | </span></span><hr /> | ||
</h2> | </h2> | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> About model </span></span | + | <span><span style="color:#7f1015"> About model </span></span> |
</h3> | </h3> | ||
| Line 452: | Line 471: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1. MiRNA is not degraded throughout the reaction process.</span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 459: | Line 478: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">2. The two fusion proteins of dCas9 and split-HRP fragments have the |
| + | |||
| + | same ability to combine with the stem-loop structure, and only when two | ||
| + | |||
| + | different proteins next to each other, can they have the ability to catalyze | ||
| + | |||
| + | substrate and produce signal.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 466: | Line 491: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">3. The number of stem-loop structures in each RCA product is equal |
| + | |||
| + | under a certain reaction time. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 473: | Line 500: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">4. The enzymatic activity remains unchanged with time under the premise |
| + | |||
| + | of excessive amount of enzymes or a short-time reaction.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 481: | Line 510: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> About the data </span></span | + | <span><span style="color:#7f1015"> About the data </span></span> |
</h3> | </h3> | ||
| Line 487: | Line 516: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1. The data we obtain from wet-lab experiment are reliable.</span> |
</p> | </p> | ||
</br> | </br> | ||
| Line 494: | Line 523: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">2. All the results are trustworthy in the process of statistical |
| + | |||
| + | processing and data calculation.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 506: | Line 537: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015">Notations </span></span | + | <span><span style="color:#7f1015">Notations </span></span> |
</h3> | </h3> | ||
| Line 512: | Line 543: | ||
<!--表格--> | <!--表格--> | ||
| − | <table class="MsoTableGrid" border="1" cellspacing="0" cellpadding="0" style="border:none;"> | + | <p> |
| − | + | <table class="MsoTableGrid" border="1" cellspacing="0" cellpadding="0" style="border:none;"> | |
| − | + | <tbody> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">Symbol </span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">Definition </span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">x</span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK7"></a><a name="OLE_LINK6"></a><span style="font-family:"">The | |
| − | + | concentration of</span><span style="font-family:""> miRNA (pM)</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">C<sub>1</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The | |
| − | + | concentration of initiated probe (Abbreviated to iprobe)</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">k<sub>1</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK8"></a><span style="font-family:"">A constant representing the scale factor</span><span style="font-family:""></span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">K<sub>m </sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">One | |
| − | + | of the characteristic constants of phi29 DNA polymerase</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">V<sub>max</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">One | |
| − | + | of the characteristic constants of phi29 DNA polymerase</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">k<sub>2</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">A | |
| − | + | constant representing the scale factor</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">V</span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The initial | |
| − | + | speed of RCA </span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK9"></a><i><span style="font-family:"">n<sub>1</sub></span></i><i><sub><span style="font-family:""></span></sub></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The | |
| − | + | moles of RCA product</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">n<sub>2</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK10"></a><span style="font-family:"">The number of stem-loop structures in each RCA product</span><span style="font-family:""></span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">n</span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The total | |
| − | + | amount of <a name="OLE_LINK23"></a><a name="OLE_LINK22"></a>stem-loop structures</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">N</span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK41"></a><a name="OLE_LINK16"></a><span style="font-family:"">The | |
| − | + | molecule number of the fusion protein of dCas9 and split-HRP fragments</span><span style="font-family:""></span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">k<sub>3</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">A | |
| − | + | constant representing the scale factor</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">y<sub>1</sub></span></i> | |
| − | DNA-dye-complex (RFU)</span> | + | </p> |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The fluorescence | |
| − | + | intensity of DNA-dye-complex (RFU)</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">N<sub>1</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The | |
| − | + | molecule number of the fusion protein of dCas9 and split-HRP fragments in the | |
| − | + | solution</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">N<sub>2</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK45"></a><span style="font-family:"">The molecule number of the fusion protein of dCas9 and | |
| − | + | split-HRP fragments binding with stem-loop structure</span><span style="font-family:""></span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">k<sub>4</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">A | |
| − | + | constant representing the scale factor</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">k<sub>5</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">A | |
| − | + | constant representing the scale factor</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">ρ</span></i><i><span style="font-family:""></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">Signal | |
| − | + | to noise ratio(Abbreviated to SNR)</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">I</span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The | |
| − | + | molecule number of formed intact HRP proteins </span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">I<sub>1</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">The | |
| − | + | molecule number of formed intact HRP proteins in the solution</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">I<sub>2</sub></span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <a name="OLE_LINK43"></a><span style="font-family:"">The molecule number of formed intact HRP proteins through | |
| − | + | binding with stem-loop structure</span><span style="font-family:""></span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">t</span></i> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <span style="font-family:"">Reaction | |
| − | + | time</span> | |
| − | + | </p> | |
| − | + | </td> | |
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | |
| − | + | <p class="MsoNormal"> | |
| − | + | <i><span style="font-family:"">y<sub>2</sub></span></i> | |
| − | </table> | + | </p> |
| − | + | </td> | |
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span style="font-family:"">The | ||
| + | signal intensity (OD<sub>450</sub>)</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </p> | ||
| Line 823: | Line 864: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The RCA model </span></span | + | <span><span style="color:#7f1015"> The RCA model </span></span> |
</h3> | </h3> | ||
| + | |||
| + | |||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, we | |
| + | |||
| + | assume that there is a linear relationship between the | ||
| + | |||
| + | concentration of the initiated probes and miRNA as the combination reaction | ||
| + | |||
| + | between miRNA and probe occurs spontaneously<sup>1 </sup> . This can be written | ||
| + | |||
| + | as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| − | |||
| Line 845: | Line 895: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, RCA is an enzymatic reaction, the reaction equation is written as: iprobe+ | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, |
| + | |||
| + | RCA is an enzymatic reaction, the reaction equation is written as: iprobe+dNTP→ | ||
| + | |||
| + | iprobe-dN+PPi, under the premise of excessive amount of enzymes and dNTPs, the | ||
| + | |||
| + | relationship between the concentration of iprobe and the initial speed of RCA | ||
| + | |||
| + | can be described by the Michaelis-Menten equation. This can be written as: | ||
| + | |||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 863: | Line 923: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">It is obvious that there is a linear relationship between the molecule numbers of iprobe-dN (n<sub>1 </sub> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">It is |
| − | , reflects the molecule numbers of RCA product) and the initial speed of RCA reaction. Notably, under the premise of excessive amount of enzymes, the extending of each RCA production is related to the reaction time, not the concentration of the iprobe. It is a linear relationship when we assume that the enzymatic activity remains unchanged with time, thus, the number of stem-loop structures (n<sub>2 </sub> | + | |
| − | ) in each RCA product is stable under the premise of a certain reaction time. </span> | + | obvious that there is a linear relationship between the molecule numbers of |
| + | |||
| + | iprobe-dN (n<sub>1 </sub> | ||
| + | , reflects the molecule numbers of RCA product) and the initial speed of RCA | ||
| + | |||
| + | reaction. Notably, under the premise of excessive amount of enzymes, the | ||
| + | |||
| + | extending of each RCA production is related to the reaction time, not the | ||
| + | |||
| + | concentration of the iprobe. It is a linear relationship when we assume that the | ||
| + | |||
| + | enzymatic activity remains unchanged with time, thus, the number of stem-loop | ||
| + | |||
| + | structures (n<sub>2 </sub> | ||
| + | ) in each RCA product is stable under the premise of a certain reaction time. | ||
| + | |||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 877: | Line 953: | ||
</br> | </br> | ||
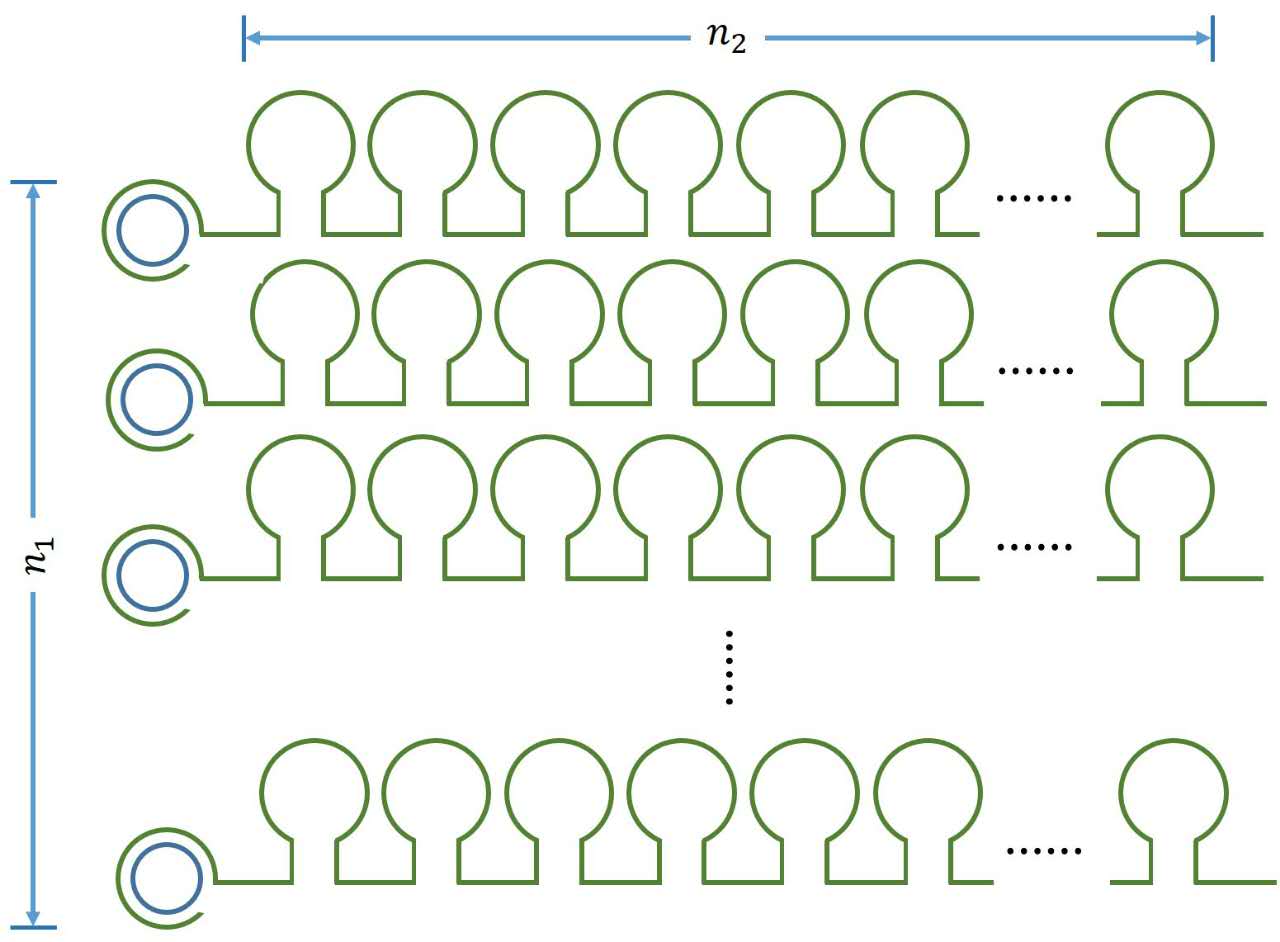
| − | + | <!--正文--> | |
| − | + | ||
| − | <!-- | + | |
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | + | ||
| + | size:18px;"><b>Figure 1. Schematic diagram</b></span> | ||
| + | </p> | ||
</br> | </br> | ||
| + | |||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This |
| + | |||
| + | can be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 914: | Line 992: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And then, SYBR Green binds to DNA. The | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And |
| − | resulting DNA-dye-complex absorbs blue light (<i>λ<sub>max</sub></i> = 497nm) and emits green light (<i>λ<sub>max</sub></i> = 520nm). Thus, there is | + | |
| − | a linear relationship between the total amount of stem-loop structures and the fluorescence | + | then, SYBR Green binds to DNA. The |
| + | resulting DNA-dye-complex absorbs blue light (<i>λ<sub>max</sub></i> = 497nm) | ||
| + | |||
| + | and emits green light (<i>λ<sub>max</sub></i> = 520nm). Thus, there is | ||
| + | a linear relationship between the total amount of stem-loop structures and the | ||
| + | |||
| + | fluorescence | ||
intensity of DNA-dye-complex. This can be written as:</span> | intensity of DNA-dye-complex. This can be written as:</span> | ||
</p> | </p> | ||
| Line 931: | Line 1,015: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In summary, the relationship between the concentration of miRNA and the fluorescence intensity of DNA-dye-complex can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In |
| + | |||
| + | summary, the relationship between the concentration of miRNA and the | ||
| + | |||
| + | fluorescence intensity of DNA-dye-complex can be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 939: | Line 1,027: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model6.jpg| | + | [[File:T--NUDT_CHINA--model6.jpg|210px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 947: | Line 1,035: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The signal detection model </span></span | + | <span><span style="color:#7f1015"> The signal detection model |
| + | |||
| + | </span></span> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, it is | |
| + | |||
| + | obvious that there is a linear relationship between | ||
| + | |||
| + | the molecule number of formed intact HRP proteins in the solution (consider as | ||
| + | |||
| + | NOISE) and the molecule number of the fusion proteins in the solution. This can | ||
| + | |||
| + | be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 969: | Line 1,067: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, we build a probability model to find the expression of the molecule number of formed intact HRP proteins (fused with dCas9) through binding with stem-loop structure (consider as SIGNAL). It is related to the total amount of stem-loop structures and the molecule number of DNA-bound fusion proteins. We obtain the result by Monte Carlo method. </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, |
| + | |||
| + | we build a probability model to find the expression of the molecule number of | ||
| + | |||
| + | formed intact HRP proteins (fused with dCas9) through binding with stem-loop | ||
| + | |||
| + | structure (consider as SIGNAL). It is related to the total amount of stem-loop | ||
| + | |||
| + | structures and the molecule number of DNA-bound fusion proteins. We obtain the | ||
| + | |||
| + | result by Monte Carlo method. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 984: | Line 1,092: | ||
</br> | </br> | ||
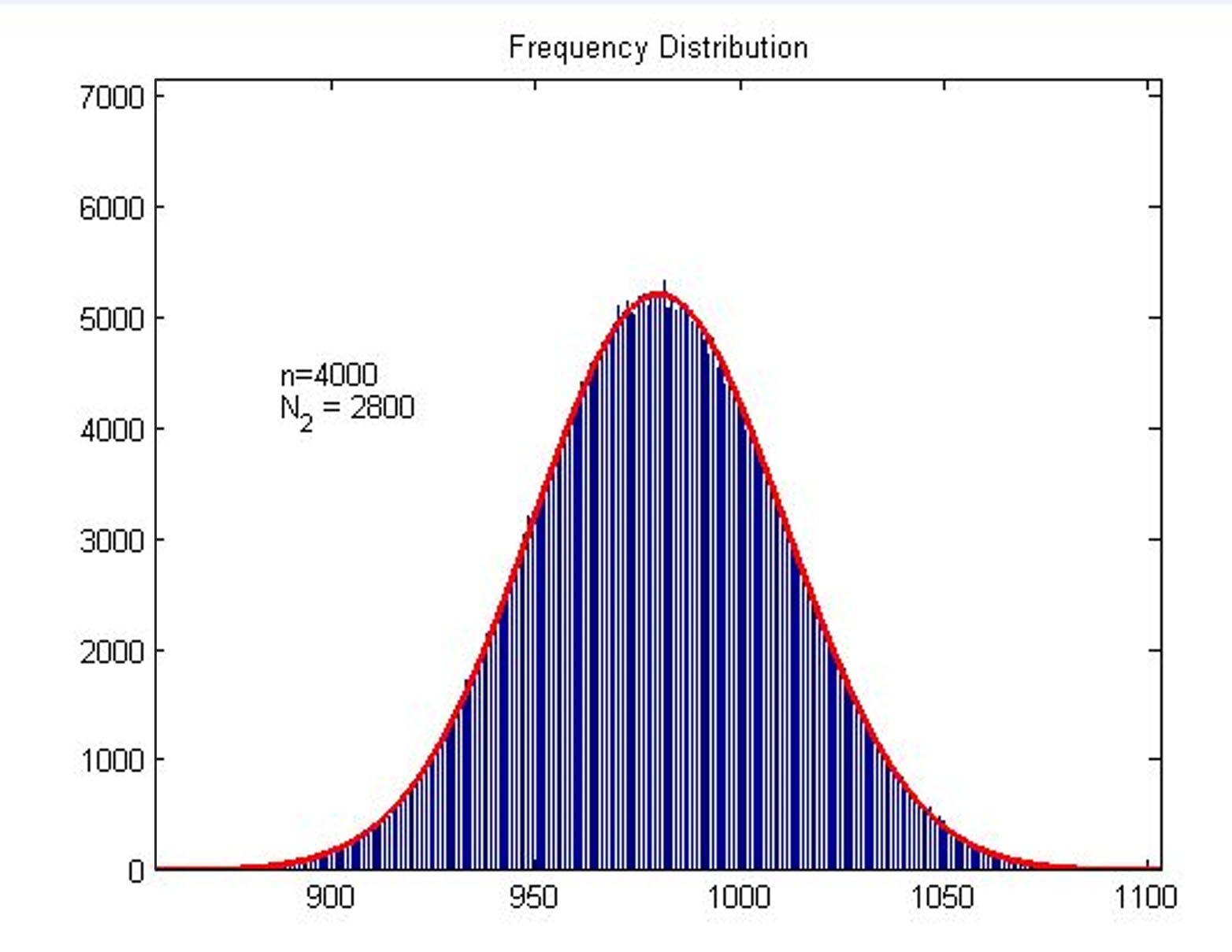
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure2. Frequency Distribution</b> Plot of frequency | ||
| + | |||
| + | distribution of the molecule number of formed intact HRP proteins through | ||
| + | |||
| + | binding with stem-loop structure. </span> | ||
</br> | </br> | ||
| Line 996: | Line 1,110: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 3. The result of the probability model</b> Plot of the | ||
| + | |||
| + | expect value of the molecule number of formed intact proteins against the total | ||
| + | |||
| + | amount of stem-loop structures, carried out at 0.1, 0.3, 0.5, 0.7, 0.9 of | ||
| + | |||
| + | N<sub>2</sub> to n ratio.</span> | ||
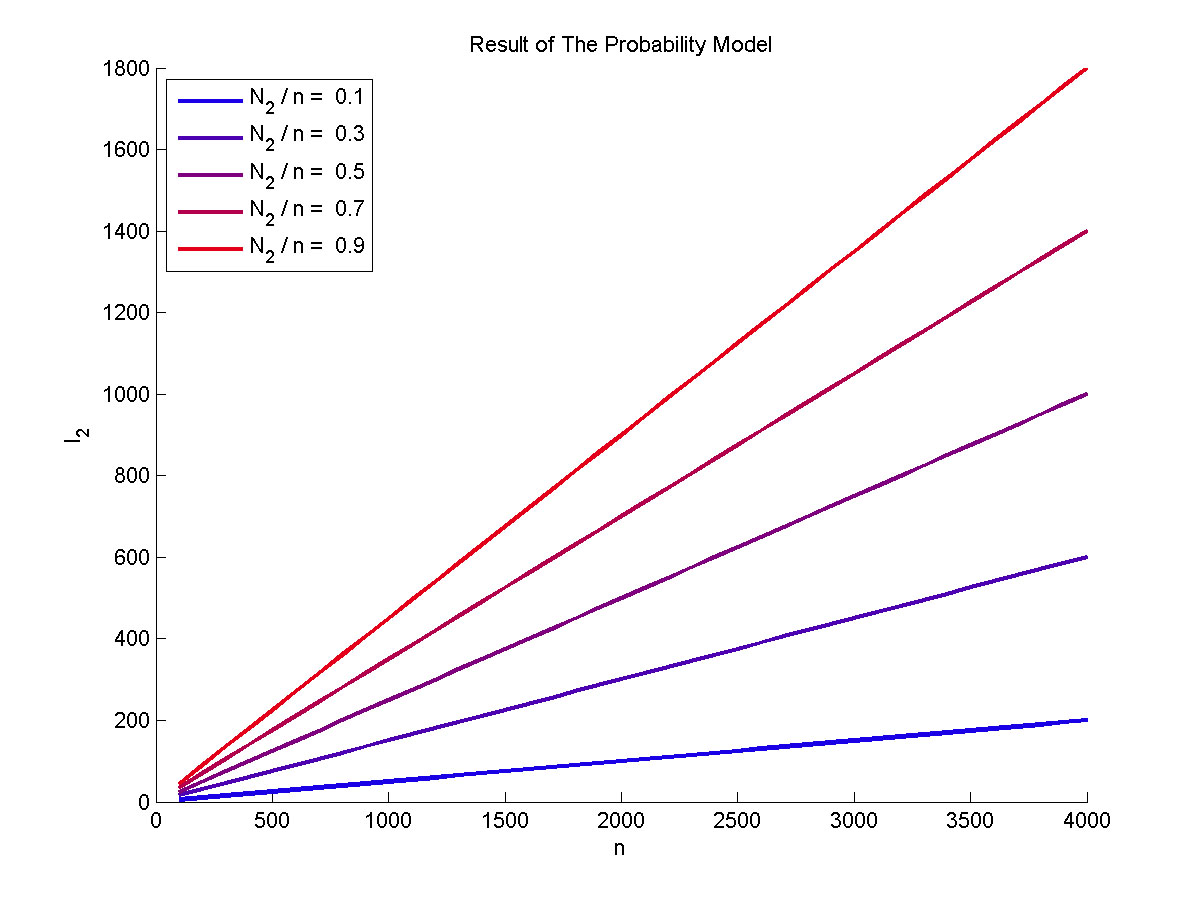
</p> | </p> | ||
</br> | </br> | ||
| Line 1,002: | Line 1,124: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The result can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | result can be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,011: | Line 1,135: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model8.jpg| | + | [[File:T--NUDT_CHINA--model8.jpg|70px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,017: | Line 1,141: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">As for <i>N<sub>2</sub></i>, | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">As for |
| + | |||
| + | <i>N<sub>2</sub></i>, | ||
which refers to the molecule number of the fusion proteins binding with | which refers to the molecule number of the fusion proteins binding with | ||
stem-loop structure. The equation is formulated based on the limiting case. The | stem-loop structure. The equation is formulated based on the limiting case. The | ||
| Line 1,029: | Line 1,155: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model9.jpg| | + | [[File:T--NUDT_CHINA--model9.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,036: | Line 1,162: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And then, we can obtain the expressions for <i>ρ</i></span> <span>and<i> I</i> separately.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And |
| + | |||
| + | then, we can obtain the expressions for <i>ρ</i></span> <span>and<i> I</i> | ||
| + | |||
| + | separately.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,045: | Line 1,175: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model10.jpg| | + | [[File:T--NUDT_CHINA--model10.jpg|50px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,058: | Line 1,188: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Notably, under | |
| + | |||
| + | the premise of excessive substrates, | ||
it is a linear relationship between the reaction rate and the concentration of | it is a linear relationship between the reaction rate and the concentration of | ||
enzymes when we assume that the enzymatic activity remain unchanged with time, | enzymes when we assume that the enzymatic activity remain unchanged with time, | ||
| − | thus, we can obtain the expression of the signal intensity at <i>t<sub>th</sub> </i>time-step. This is | + | thus, we can obtain the expression of the signal intensity at <i>t<sub>th</sub> |
| + | |||
| + | </i>time-step. This is | ||
written as:</span> | written as:</span> | ||
</p> | </p> | ||
| Line 1,071: | Line 1,205: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model12.jpg| | + | [[File:T--NUDT_CHINA--model12.jpg|70px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,079: | Line 1,213: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In summary, the relationship between the number of stem-loop structures and SNR under the premise of adding a certain amount of the fusion proteins can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In |
| + | |||
| + | summary, the relationship between the number of stem-loop structures and SNR | ||
| + | |||
| + | under the premise of adding a certain amount of the fusion proteins can be | ||
| + | |||
| + | written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,096: | Line 1,236: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The relationship between the number of Stem-loop structures and the signal intensity under the premise of adding a certain amount of the fusion proteins can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | relationship between the number of Stem-loop structures and the signal intensity | ||
| + | |||
| + | under the premise of adding a certain amount of the fusion proteins can be | ||
| + | |||
| + | written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,105: | Line 1,251: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model14.jpg| | + | [[File:T--NUDT_CHINA--model14.jpg|280px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,111: | Line 1,257: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The calculation of the constants </span></span | + | <span><span style="color:#7f1015"> The calculation of the constants |
| + | |||
| + | </span></span> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To simplify the equation (1), the constants | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To |
| − | can be integrated. As can be seen from the above table, <i>k<sub>1</sub></i>, <i>k<sub>2</sub></i>, <i>k<sub>3</sub></i><sub> </sub>are | + | |
| − | constants representing the scale factor, <i>K<sub>m</sub></i> and <i>V<sub>max</sub></i> are | + | simplify the equation (1), the constants |
| − | characteristic constants of phi 29 DNA polymerase, and <i>n<sub>2</sub></i>, which refers to the number of stem-loop structures | + | can be integrated. As can be seen from the above table, <i>k<sub>1</sub></i>, |
| + | |||
| + | <i>k<sub>2</sub></i>, <i>k<sub>3</sub></i><sub> </sub>are | ||
| + | constants representing the scale factor, <i>K<sub>m</sub></i> and | ||
| + | |||
| + | <i>V<sub>max</sub></i> are | ||
| + | characteristic constants of phi 29 DNA polymerase, and <i>n<sub>2</sub></i>, | ||
| + | |||
| + | which refers to the number of stem-loop structures | ||
in each RCA product, is stable under the premise of a certain reaction time. | in each RCA product, is stable under the premise of a certain reaction time. | ||
| − | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the equation can be | + | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the equation |
| + | |||
| + | can be | ||
simplified as:</span> | simplified as:</span> | ||
</p> | </p> | ||
| Line 1,129: | Line 1,287: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model15.jpg| | + | [[File:T--NUDT_CHINA--model15.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,135: | Line 1,293: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
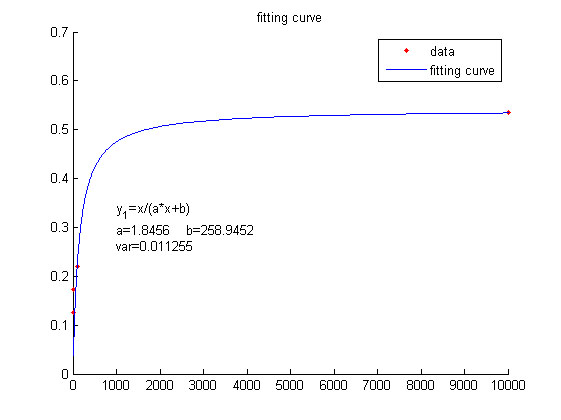
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We use this equation to fit the data points obtained through experiments. (Figure | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We use |
| + | |||
| + | this equation to fit the data points obtained through experiments. (Figure 2. | ||
| + | |||
| + | (D)in the <a href="/Team:NUDT_CHINA/Results">RESULTS.page</a>) The fitting | ||
| + | |||
| + | curve is shown below.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,147: | Line 1,311: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 4. Fitting curve</b> Fitting curve of fluorescence | ||
| + | |||
| + | intensity of DNA-dye-complex (RFU) against the concentrations of miRNA | ||
| + | |||
| + | (pM).</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,154: | Line 1,324: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">With regard to the equation (2), we set the parameters based on the experiments as follows: | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">With |
| − | <i>k<sub>3</sub> </i>=8.35*10<sup>-16 </sup>(<i>n</i>=<i>y<sub>1</sub></i>/<i>k<sub>3</sub></i>); <i>k<sub>4</sub></i> =0.038;<i> k<sub>5</sub>t</i>=5.9*10<sup>-12</sup>. | + | |
| + | regard to the equation (2), we set the parameters based on the experiments as | ||
| + | |||
| + | follows: | ||
| + | <i>k<sub>3</sub> </i>=8.35*10<sup>-16 </sup> | ||
| + | |||
| + | (<i>n</i>=<i>y<sub>1</sub></i>/<i>k<sub>3</sub></i>); <i>k<sub>4</sub></i> | ||
| + | |||
| + | =0.038;<i> k<sub>5</sub>t</i>=5.9*10<sup>-12</sup>. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 1,163: | Line 1,341: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> Results and Analysis </span></span | + | <span><span style="color:#7f1015"> Results and Analysis |
| + | |||
| + | </span></span> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By integrating the two models, we can obtain the relationships between SNR, the signal intensity respectively and the concentration of miRNA under different addition amount of fusion proteins. | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By |
| + | |||
| + | integrating the two models, we can obtain the relationships between SNR, the | ||
| + | |||
| + | signal intensity respectively and the concentration of miRNA under different | ||
| + | |||
| + | addition amount of fusion proteins. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 1,175: | Line 1,361: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The results are shown below.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | results are shown below.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,186: | Line 1,374: | ||
</br> | </br> | ||
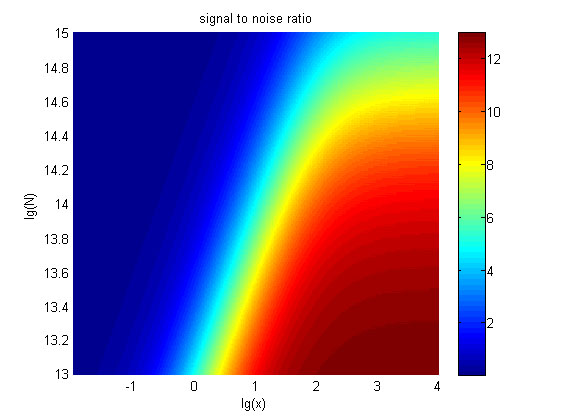
<p> | <p> | ||
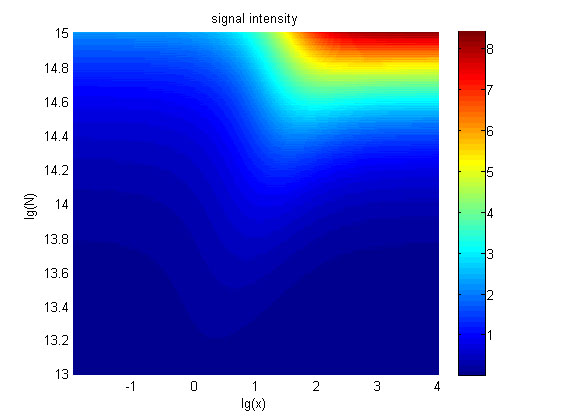
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | 5. The signal intensity (OD<sub>450</sub>) Three-dimensional map of signal intensity (OD<sub>450</sub>) | + | |
| − | against miRNA concentration(pM) and additional amount of fusion proteins.</span | + | size:18px;"><b>Figure |
| + | 5. The signal intensity (OD<sub>450</sub>)</b> Three-dimensional map of signal | ||
| + | |||
| + | intensity (OD<sub>450</sub>) | ||
| + | against miRNA concentration(pM) and additional amount of fusion proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,194: | Line 1,386: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">There exists a monotonous relation between the concentration of miRNA and the signal intensity when the value of the molecule number of the fusion proteins is relatively large. While the relationship does not hold when the value of the molecule number of the fusion proteins is relatively small. And the signal intensity increases as the value of the molecule number of the fusion proteins increases.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">There |
| + | |||
| + | exists a monotonous relation between the concentration of miRNA and the signal | ||
| + | |||
| + | intensity when the value of the molecule number of the fusion proteins is | ||
| + | |||
| + | relatively large. While the relationship does not hold when the value of the | ||
| + | |||
| + | molecule number of the fusion proteins is relatively small. And the signal | ||
| + | |||
| + | intensity increases as the value of the molecule number of the fusion proteins | ||
| + | |||
| + | increases.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig6.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;"><b>Figure 6. The result of SNR</b> Three-dimensional map of signal | ||
| + | |||
| + | to noise ratio against miRNA concentration (pM) and additional amount of fusion | ||
| + | |||
| + | proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,201: | Line 1,424: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | signal-to-noise ratio decreases as the value of the molecule number of fusion | ||
| + | |||
| + | proteins increases. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> Test of model | ||
| + | |||
| + | </span></span> | ||
| + | </h3> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Our | ||
| + | |||
| + | model predicts the relationship between the signal intensity and the | ||
| + | |||
| + | concentration of miRNA under the premise of adding a certain amount of the | ||
| + | |||
| + | fusion proteins of dCas9 and split-HRP fragments. So we can test if our model is | ||
| + | |||
| + | reasonable by comparing the curves predicted by the model with those obtained | ||
| + | |||
| + | from the experiment. The results are shown below. | ||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| Line 1,209: | Line 1,462: | ||
</br> | </br> | ||
</html> | </html> | ||
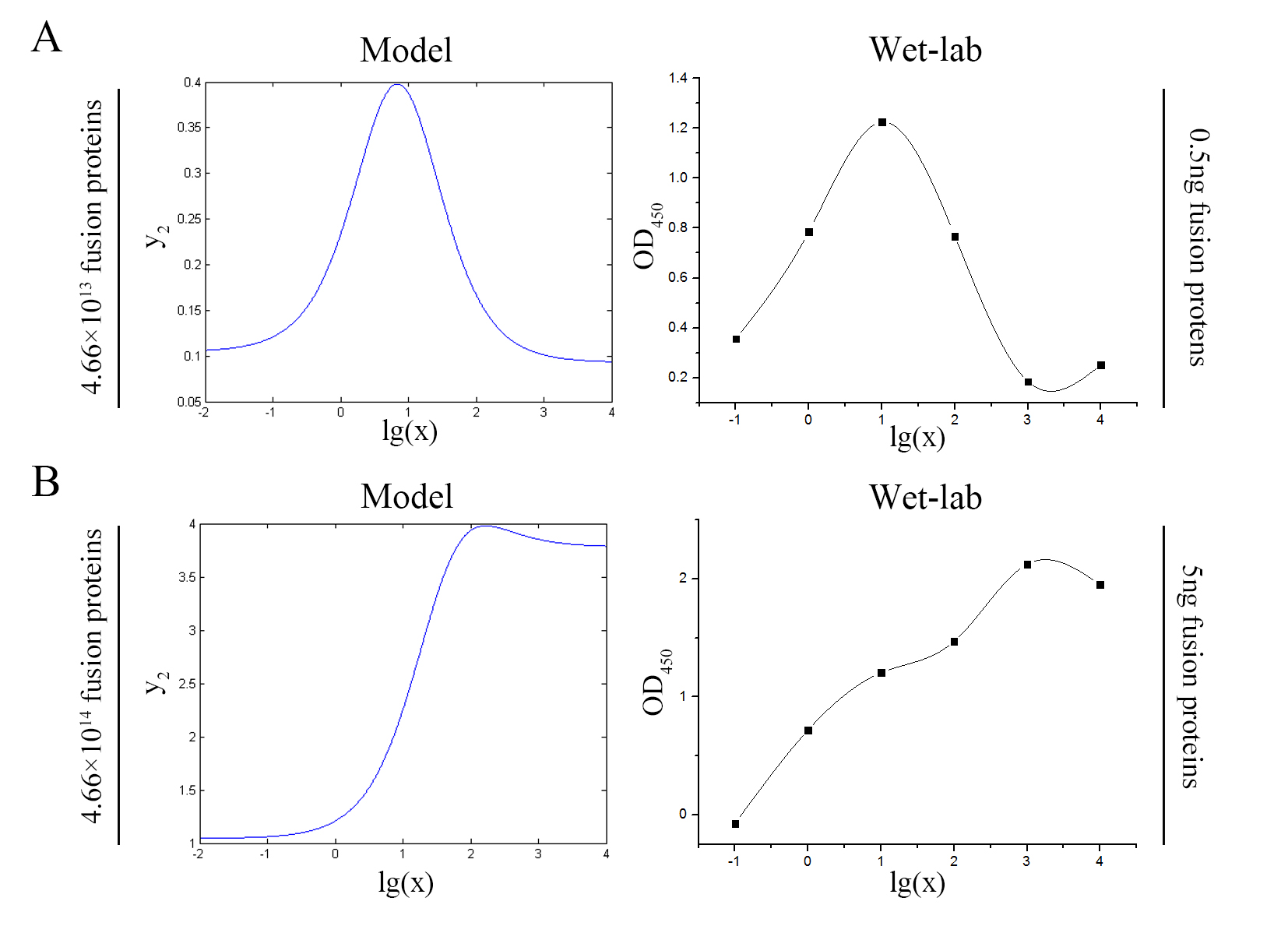
| − | [[File:T--NUDT_CHINA-- | + | [[File:T--NUDT_CHINA--modelfig7-2.jpg|700px|center]] |
<html> | <html> | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 7. Test of model</b> Plot of the signal intensity (OD<sub>450</sub>) against miRNA concentration (pM) predicted by the model and experimented by wet-lab. (A) The value of the molecule number of each fusion protein added to the system is set to be 4.66e+13. The mass of each fusion protein added to the system in wet-lab is 0.5 ng. (B) The value of the molecule number of each fusion protein added to the system is set to be 4.66e+14. The mass of each fusion protein added to the system in wet-lab is 5 ng.</span> | ||
</p> | </p> | ||
| + | |||
</br> | </br> | ||
| + | |||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We can learn | |
| + | |||
| + | from the figure above that the general trend of curves predicted by the model agree well with the experimental results. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> Model extension | ||
| + | |||
| + | </span></span> | ||
| + | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Furthermore, we | |
| + | |||
| + | consider that it is conducive to signal detection when the value of signal | ||
| + | |||
| + | intensity and SNR are both greater than two. We obtain the ideal region through | ||
| + | |||
| + | calculation, which is shown as below. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
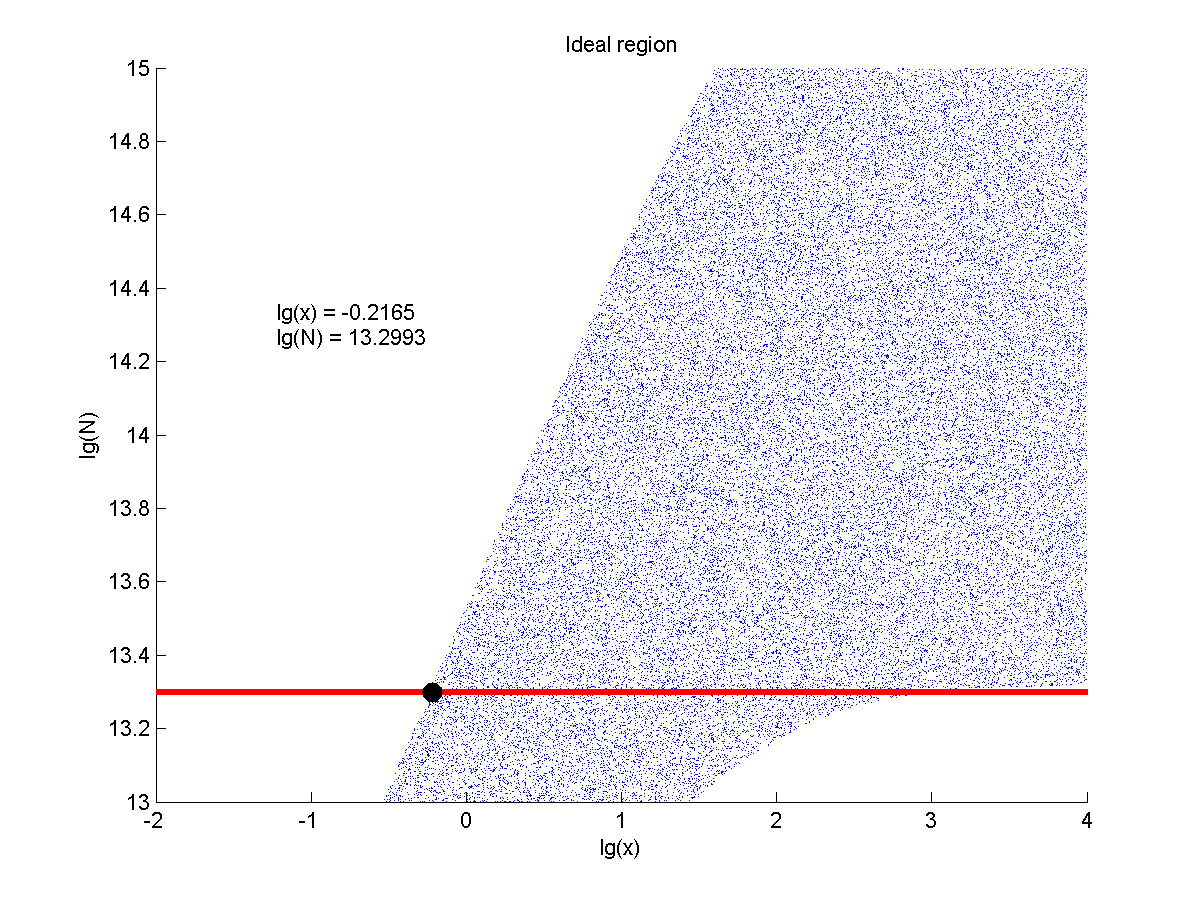
| Line 1,240: | Line 1,513: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 8. Ideal region</b> Region of qualified logarithm of | ||
| + | |||
| + | concentrations of miRNA (pM) and logarithm of the additional amount of fusion | ||
| + | |||
| + | proteins.</span> | ||
</p> | </p> | ||
| Line 1,249: | Line 1,528: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">When the value of N is set to | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">When |
| + | |||
| + | the value of N is set to e+13.3, the range of the value of x contained in the | ||
| + | |||
| + | ideal region is a maximum, which means when the value of the molecule number of | ||
| + | |||
| + | the fusion proteins is e+13.3, the concentration range of miRNA that can be | ||
| + | |||
| + | detected is maximum. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,261: | Line 1,548: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Based on our simulation, we came to the conclusion that:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Based |
| + | |||
| + | on our simulation, we came to the conclusion that:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,268: | Line 1,557: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The general trend of curves predicted by the model are in good agreement with the experimental |
| + | |||
| + | results, thus indicating that our model is reasonable.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,274: | Line 1,565: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The value of the molecule number of fusion protein of dCas9 and split-HRP fragments not only affects the signal-to-noise ratio, but also the signal intensity. So we need to weigh its impact on both to select the optimal solution.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | value of the molecule number of the fusion protein of dCas9 and split-HRP | ||
| + | |||
| + | fragments not only affects the signal-to-noise ratio, but also the signal | ||
| + | |||
| + | intensity. So we need to weigh its impact on both to select the optimal | ||
| + | |||
| + | solution.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,281: | Line 1,580: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The concentration range of miRNA | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | concentration range of miRNA that can be detected is a maximum when we set the | ||
| + | |||
| + | value of the molecule number of the fusion proteins to be e+13.3, building on | ||
| + | |||
| + | which, our wet-lab protocol could be optimized in our future work.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,292: | Line 1,597: | ||
<!--正文--> | <!--正文--> | ||
| − | <p | + | <p > |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1 Deng, R. <i>et al</i>.Toehold-initiated rolling circleamplification for visualizing individual microRNAs in situ in single cells. <i>Angew Chem Int Ed Engl </i>53, 2389-2393,doi:10.1002/anie.201309388 (2014).</span> | |
| − | + | ||
| − | doi:10.1002/anie.201309388 (2014).</span> | + | |
</p> | </p> | ||
</br> | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 1,313: | Line 1,635: | ||
</div> | </div> | ||
<!-- end #content-wrap --> | <!-- end #content-wrap --> | ||
| − | + | <div style="border:solid 1px rgb(10,31,84);background-color:rgb(10,31,84);"> | |
| − | + | <div class="row footer-link" style="border:solid 0px red;margin-top:100px;"> | |
| − | + | <style>.menu-link-container ul li{ list-style:none; } .menu-link-container a{ color:white; font-size:14px;line-height:26px; } | |
| − | + | .menu-link-container a:hover{ | |
| − | + | text-decoration:underline; | |
| − | + | } #c_menu9909 .medium-6{ text-align:center; width:auto; border:solid 0px red; } #c_menu9909 .medium-6 h5{color:white;}#c_menu9909 .medium-6{height:315px;border:solid 0px red;}</style> | |
| − | + | <div id="c_menu9909"> | |
| − | + | <!--div class="medium-6 columns" style="width:auto;"> | |
| − | + | <h5 class="column-header">HOME</h5> | |
| − | + | <div class="menu-link-container"></div></div--> | |
| − | + | <div class="medium-6 columns" style="width:auto;"> | |
| − | + | <h5 class="column-header">TEAM</h5> | |
| − | + | <div class="menu-link-container"> | |
| − | + | ||
| − | + | ||
| − | + | <a href="/Team:NUDT_CHINA/Team">Team</a><br/> | |
| − | + | ||
| − | + | <a href="/Team:NUDT_CHINA/Collaborations">Collaborations</a><br/> | |
| − | + | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">PROJECT</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Description">Description</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Design">Design</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Experiments">Experiments</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Proof">Proof of Concept</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Demonstrate">Demonstrate</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Results">Results</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Notebook">Notebook</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">PARTS</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Parts">Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Basic_Part">Basic Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Composite_Part">Composite Parts</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Part_Collection">Part Collection</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <a href="/Team:NUDT_CHINA/Safety" style="text-decoration:none;"> | ||
| + | <h5 class="column-header">SAFETY</h5></a> | ||
| + | |||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <a href="/Team:NUDT_CHINA/Attributions" style="text-decoration:none;"> | ||
| + | <h5 class="column-header">ATTRIBUTIONS</h5></a> | ||
| + | |||
| + | </div> | ||
| + | <!--div class="medium-6 columns" style="width:auto;"> | ||
| + | <A href="/Team:NUDT_CHINA/Attributions" style="text-decoration:none;"><h5 class="column-header">ATTRIBUTIONS</h5></A> | ||
| + | <div class="menu-link-container"></div></div--> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">HUMAN | ||
| + | <br>PRACTICES</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Human_Practices">Human Practices</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/HP/Silver">Silver</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/HP/Gold">Gold</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Integrated_Practices">Integrated Practices</a><br/> | ||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Engagement">Engagement</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:auto;"> | ||
| + | <h5 class="column-header">AWARDS</h5> | ||
| + | <div class="menu-link-container"> | ||
| + | |||
| + | |||
| + | <a href="/Team:NUDT_CHINA/Model">Model</a><br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <div class="medium-6 columns" style="width:0px;height:0px;"></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
<footer id="footer"> | <footer id="footer"> | ||
<div class="row footer-link"> | <div class="row footer-link"> | ||
| Line 1,463: | Line 1,786: | ||
--> | --> | ||
| − | + | <!-- to top edit by inksci---> | |
| + | <style> | ||
| + | #to_top{ | ||
| + | position:fixed; | ||
| + | bottom:100px; | ||
| + | right:0px; | ||
| + | width:60px; | ||
| + | height:60px; | ||
| + | font-size:14px; | ||
| + | line-height:60px; | ||
| + | text-align:center; | ||
| + | background-color:black; | ||
| + | color:rgb(246,168,0); | ||
| + | cursor:pointer; | ||
| + | } | ||
| + | </style> | ||
| + | <div onclick="top_it()" id="to_top"> | ||
| + | TOP | ||
| + | </div> | ||
| + | <script type="text/javascript"> | ||
| + | var oTop = document.getElementById("to_top"); | ||
| + | oTop.style.display="none"; | ||
| + | var top_break=false; | ||
| + | /** | ||
| + | * 回到页面顶部 | ||
| + | * @param acceleration 加速度 | ||
| + | * @param time 时间间隔 (毫秒) | ||
| + | **/ | ||
| + | var save_scrolltop=100000000000000; | ||
| + | window.onscroll = function(){ | ||
| + | |||
| + | |||
| + | var scrolltop = document.documentElement.scrollTop || document.body.scrollTop; | ||
| + | if(scrolltop>300){ | ||
| + | oTop.style.display="block"; | ||
| + | }else{ | ||
| + | oTop.style.display="none"; | ||
| + | } | ||
| + | if(scrolltop>save_scrolltop) | ||
| + | { | ||
| + | top_break=true; | ||
| + | } | ||
| + | save_scrolltop=scrolltop; | ||
| + | } | ||
| + | function top_it(){ | ||
| + | top_break=false; | ||
| + | goTop();return false; | ||
| + | } | ||
| + | function goTop(acceleration, time) { | ||
| + | if(top_break){ | ||
| + | return; | ||
| + | } | ||
| + | oTop.style.display="none"; | ||
| + | acceleration = acceleration || 0.1; | ||
| + | time = time || 16; | ||
| + | |||
| + | var x1 = 0; | ||
| + | var y1 = 0; | ||
| + | var x2 = 0; | ||
| + | var y2 = 0; | ||
| + | var x3 = 0; | ||
| + | var y3 = 0; | ||
| + | |||
| + | if (document.documentElement) { | ||
| + | x1 = document.documentElement.scrollLeft || 0; | ||
| + | y1 = document.documentElement.scrollTop || 0; | ||
| + | } | ||
| + | if (document.body) { | ||
| + | x2 = document.body.scrollLeft || 0; | ||
| + | y2 = document.body.scrollTop || 0; | ||
| + | } | ||
| + | var x3 = window.scrollX || 0; | ||
| + | var y3 = window.scrollY || 0; | ||
| + | |||
| + | // 滚动条到页面顶部的水平距离 | ||
| + | var x = Math.max(x1, Math.max(x2, x3)); | ||
| + | // 滚动条到页面顶部的垂直距离 | ||
| + | var y = Math.max(y1, Math.max(y2, y3)); | ||
| + | |||
| + | // 滚动距离 = 目前距离 / 速度, 因为距离原来越小, 速度是大于 1 的数, 所以滚动距离会越来越小 | ||
| + | var speed = 1 + acceleration; | ||
| + | window.scrollTo(Math.floor(x / speed), Math.floor(y / speed)); | ||
| + | |||
| + | // 如果距离不为零, 继续调用迭代本函数 | ||
| + | if(x > 0 || y > 0) { | ||
| + | var invokeFunction = "goTop(" + acceleration + ", " + time + ")"; | ||
| + | window.setTimeout(invokeFunction, time); | ||
| + | } | ||
| + | } | ||
| + | </script> | ||
| + | <!-- to top edit by inksci---> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 13:34, 19 October 2016
TOP