F1yingFish (Talk | contribs) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 7,294: | Line 7,294: | ||
The use of multiple lists further enhances the advantage of this modular system over de novo synthesis because it means we can use the same blocks make times over. </p> | The use of multiple lists further enhances the advantage of this modular system over de novo synthesis because it means we can use the same blocks make times over. </p> | ||
<p class="lead">For our proof of concept we have chosen to encode words. This is conceptually the easiest encoding of data to follow. We wish to encode large bits of data, by adding together smaller, modular chunks. Sentences (BabbleBlocks) are large bits of data made up of words (BabbleBricks). Anything however can be encoded in this region; numbers for chunks of binary code, pixel colours for pictures, musical notes to encode a song</p> | <p class="lead">For our proof of concept we have chosen to encode words. This is conceptually the easiest encoding of data to follow. We wish to encode large bits of data, by adding together smaller, modular chunks. Sentences (BabbleBlocks) are large bits of data made up of words (BabbleBricks). Anything however can be encoded in this region; numbers for chunks of binary code, pixel colours for pictures, musical notes to encode a song</p> | ||
| − | <p class="lead">Stop Region: This 17 base pair long sequence introduces a stop codon into every reading frame within a BabbleBrick. This helps prevent large BabbleBrick constructs from producing any protein when replicating in cells. Read more about why we chose to include these here.</p> | + | <p class="lead">Stop Region: This 17 base pair long sequence introduces a stop codon into every reading frame within a BabbleBrick. This helps prevent large BabbleBrick constructs from producing any protein when replicating in cells. Read more about why we chose to include these <a href="https://2016.igem.org/Team:Edinburgh_UG/Safety/Biological_Safety" target="_self">here</a>.</p> |
| − | <p class="lead">ORC Region: The ORC is a vector of error-correcting code intrinsic to each BabbleBrick. This helps rectify any errors introduced by mutation or poor-sequencing in the word coding region with high fidelity. Like the word coding region, it is staggered with nucleotides to allow fine control of the overall sequence. Read more about how an ORC works and our error-correcting functions here.</p> | + | <p class="lead">ORC Region: The ORC is a vector of error-correcting code intrinsic to each BabbleBrick. This helps rectify any errors introduced by mutation or poor-sequencing in the word coding region with high fidelity. Like the word coding region, it is staggered with nucleotides to allow fine control of the overall sequence. Read more about how an ORC works and our error-correcting functions <a href="https://2016.igem.org/Team:Edinburgh_UG/Error_Correction" target="_self">here</a>.</p> |
<p class="lead">Flanking each BabbleBrick are hangs, which are used for assembly. There are two types of hangs; A and B. Each BabbleBrick is present in two forms, an AB and a BA form. | <p class="lead">Flanking each BabbleBrick are hangs, which are used for assembly. There are two types of hangs; A and B. Each BabbleBrick is present in two forms, an AB and a BA form. | ||
In the AB BabbleBrick above.</p> | In the AB BabbleBrick above.</p> | ||
| Line 7,304: | Line 7,304: | ||
<div class="featurette" id="about"> | <div class="featurette" id="about"> | ||
| − | <h2 class="featurette-heading"> | + | <h2 class="featurette-heading">BabbleBlock Format |
</h2> | </h2> | ||
<p class="lead">The assembly method is our proposed standard for text assembly. It is based off a 2009 iGEM project. The Alberta iGEM team created a new assembly method for gene assembly. This was further improved by their 2010 team. </p> | <p class="lead">The assembly method is our proposed standard for text assembly. It is based off a 2009 iGEM project. The Alberta iGEM team created a new assembly method for gene assembly. This was further improved by their 2010 team. </p> | ||
Latest revision as of 00:59, 20 October 2016
Design
BabbleBrick Format
We went through an iterative design process with our BabbleBricks, which we have continued up to the Jamboree and will follow through on afterwards. Version 1.5 of the BabbleBrick is the standard we largely used in our proof of concept and summer work, and is the standard we submitted to the registry.
Key considerations we had throughout the design process included:
- The length of a BabbleBrick. Too long would reduce the density of information, and be more difficult to insert BabbleBlocks into a plasmid. Too short however would be difficult to work with in the lab. We originally wanted to use very short bits of DNA (~10bp) but this was too short to work with. Having to make each BabbleBrick longer however benefitted us because we now have space to include features such as error correction.
- Inclusion of stop codons. Discussion with various people resulted in the decision to add a stop codon region. This is both to help the cell by ensuring it is not unnecessarily making protein, and as a safety feature.
- The types of hangs to be used. We considered utilising enzymes that only work on methylated DNA to ensure that we only had addition of one BabbleBrick at a time. This method however was discarded and replaced with the current system of A and B hangs.
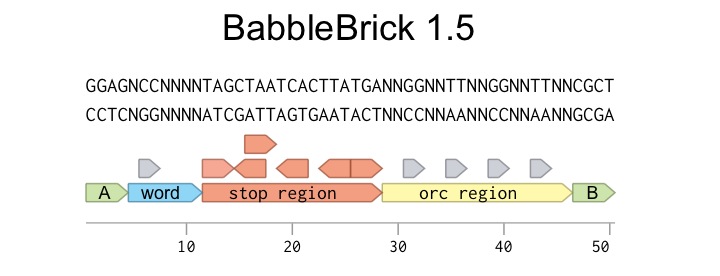
Every BabbleBrick 1.5 has the format you can see here.
The main body of the BabbleBrick consists of three regions; Word coding, Stop, and ORC
Word Coding Region: This region is 7bp long, 5 of which are unique to each BabbleBrick. The other two bases are spacers which prevent illegal restriction sites. There are 1024 different possible combinations of 5bases, and so we can encode 1024 different words in BabbleBricks. We have chosen 5bp for simplicity, however this can be extrapolated to 6 or 7 bases, which would give many more combinations.
The 1024 combinations are arbitrarily assigned to the data. A list of data segments, eg words, and the list of 5 base combinations are mapped together. One advantage of this design means that different libraries can be used.

The coding sequence: ATCGG-GAGTC-GGTCA-GTAC can be made. If the computer is instructed to decode using list 1 it will read “Team Edinburgh love pizza”. If instructed to decode using list 2 it will read “iGEM is really great”.
The instructions to use a certain decoding list can be in the address. The use of multiple lists further enhances the advantage of this modular system over de novo synthesis because it means we can use the same blocks make times over.
For our proof of concept we have chosen to encode words. This is conceptually the easiest encoding of data to follow. We wish to encode large bits of data, by adding together smaller, modular chunks. Sentences (BabbleBlocks) are large bits of data made up of words (BabbleBricks). Anything however can be encoded in this region; numbers for chunks of binary code, pixel colours for pictures, musical notes to encode a song
Stop Region: This 17 base pair long sequence introduces a stop codon into every reading frame within a BabbleBrick. This helps prevent large BabbleBrick constructs from producing any protein when replicating in cells. Read more about why we chose to include these here.
ORC Region: The ORC is a vector of error-correcting code intrinsic to each BabbleBrick. This helps rectify any errors introduced by mutation or poor-sequencing in the word coding region with high fidelity. Like the word coding region, it is staggered with nucleotides to allow fine control of the overall sequence. Read more about how an ORC works and our error-correcting functions here.
Flanking each BabbleBrick are hangs, which are used for assembly. There are two types of hangs; A and B. Each BabbleBrick is present in two forms, an AB and a BA form. In the AB BabbleBrick above.
A: The 5’ A type hang is produced upon digesting an AB type BabbleBrick with BsaI. It can anneal to the 3’ A hang of BA parts.
B: The 3’ ‘B’ type hang is produced upon digesting a AB type BabbleBrick with BsaI. It can anneal to the 5’ B hang of BA parts.
BabbleBricks are housed in plasmids as phytobricks, and are excised as above using BsaI. BabbleBlocks are constructs of multiple BabbleBricks, which can then again be housed in phytobricks.
BabbleBlock Format
The assembly method is our proposed standard for text assembly. It is based off a 2009 iGEM project. The Alberta iGEM team created a new assembly method for gene assembly. This was further improved by their 2010 team.
We have adapted their method for our assembly of much smaller chunks of DNA.
The key feature of this assembly process is the overhangs. A overhangs can only bind to other A overhangs, B overhangs can only bind to B overhangs. As we see below BabbleBricks are either 3’A 5’B or 3’B 5’A. We call these AB and BA BabbleBricks. To assemble these we must alternate the type of BabbleBrick used; AB-BA-AB-BA-AB-BA. To do this we have two libraries of every word, an AB library and a BA library.

BabbleBlocks are assembled on a magnetic bead which has a polyT tail.

An anchor piece of DNA is added to this BabbleBlock. This has a single stranded polyA stretch, which base pairs with the polyT tail. The anchor also has an A overhang, which allows addition of the first word.

An important aspect of this anchor-bead interaction is the two TG residues after the polyA section. These are spacer residues and ensure that he bead does not get ligated to the anchor. This is important for the final assembly step when the DNA construct is removed from the bead. The anchor and bead are only held together by this base pairing interaction. The length of this overlap we decided by looking at the melting temperature of different overlap lengths.
The A overhang on this construct allows the first word to be ligated on using ligase. This word must be an AB word. After this addition, a wash step occurs, which removes all excess of word 1. The magnetic bead allows the growing construct to be immobilised during this wash. Word 2, and AB word can then be added, and again a wash step follows. This continues until the desired construct (eg. A sentence) has been made.

Once the sentence has been constructed the checksum is added. This is 4 BabbleBricks which are numbers. To find out how we determine the checksum click here. The checksum detects errors in the coding regions.
The next important segment of the BabbleBlock is the address. This BabbleBrick encodes information on where the BabbleBlock is in regards to other BabbleBlocks. For example this could be:
- Chapter 3, paragraph 2, line 4 of a book
- Line 1 of a row of pixels in a picture.
The address also includes information about which data list (or lexicon) was used to encode the information.
After these BabbleBricks have been added, the final thing to add is the terminator. The 5’ end of the terminator is an A or B overhang, which allows it to ligate to the end of the checksum BabbleBricks.

Once this construct is complete with terminator it is removed from the bead. It can simply be melted off because the anchor-bead interactions are base pair interactions, not ligation of DNA.
We are then left with a linear construct, or device. This can then be stored as is, or ligated into a plasmid. To insert into a plasmid we utilise features of the anchor and terminator. These both have BsmBI cut sites, which will produce compatible overhangs for insertion of construct into a phytobrick.





