Qiuxinyuan12 (Talk | contribs) |
Qiuxinyuan12 (Talk | contribs) |
||
| Line 384: | Line 384: | ||
<div style="margin-top:80px;" id="content-wrap-ink"><!-- adjust in add_JS --> | <div style="margin-top:80px;" id="content-wrap-ink"><!-- adjust in add_JS --> | ||
<div class="row footer-link" style=""> | <div class="row footer-link" style=""> | ||
| − | |||
<h2> | <h2> | ||
| − | <span>Abstract</span> | + | <span><span style="color:#7f1015">Abstract</span></span><hr /> |
</h2> | </h2> | ||
| − | <p | + | |
| − | + | <p style="text-indent:22pt;"> | |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">MicroRNAs, | |
| − | + | serve as critical gene expression regulators at the transcriptional and | |
| − | <span style="font-size:18px;">MicroRNAs, serve as critical gene | + | post-transcriptional levels, have also been found as important blood-based |
| − | expression regulators at the transcriptional and post-transcriptional levels, | + | biomarkers for early detection of cancers. However, their current in vitro |
| − | have also been found as important blood-based biomarkers for early detection of | + | detection methods are relatively complex, costly and low sensitive. Our project |
| − | cancers. However, their current in vitro detection methods are relatively | + | attempts to establish a novel in vitro microRNA detection system which is |
| − | complex, costly and low sensitive. Our project attempts to establish a novel in | + | rapid, efficient, sensitive and specific. In this system, CRISPR-Cas9 technique |
| − | vitro microRNA detection system which is rapid, efficient, sensitive and | + | is modified to integrate with split-luciferase or split-HRP reporting systems. |
| − | specific. In this system, CRISPR-Cas9 technique is modified to integrate with | + | The advanced rolling circle amplification technology and cell-free expression |
| − | split-luciferase or split-HRP reporting systems. The advanced rolling circle | + | system are also involved and optimized. This system may ideally be compatible |
| − | amplification technology and cell-free expression system are also involved and | + | for the detection of various series of small non-coding RNAs. To our knowledge, |
| − | optimized. This system may ideally be compatible for the detection of various | + | we are the first to use the CRISPR-Cas9 system as a small non-coding RNA |
| − | series of small non-coding RNAs. To our knowledge, we are the first to use the | + | monitor in vitro. Its establishment and further development might provide a new |
| − | CRISPR-Cas9 system as a small non-coding RNA monitor in vitro. Its | + | approach for rapid and low-cost cancer screening, virus detection and curative |
| − | establishment and further development might provide a new approach for rapid | + | efficacy assessment.</span> |
| − | and low-cost cancer screening, virus detection and curative efficacy | + | |
| − | assessment. | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
| + | </br> | ||
<h2> | <h2> | ||
| − | + | <span><span style="color:#7f1015">Introduction</span></span><hr /> | |
</h2> | </h2> | ||
| − | <p> | + | <p style="text-indent:22pt;"> |
| − | < | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Nowadays, |
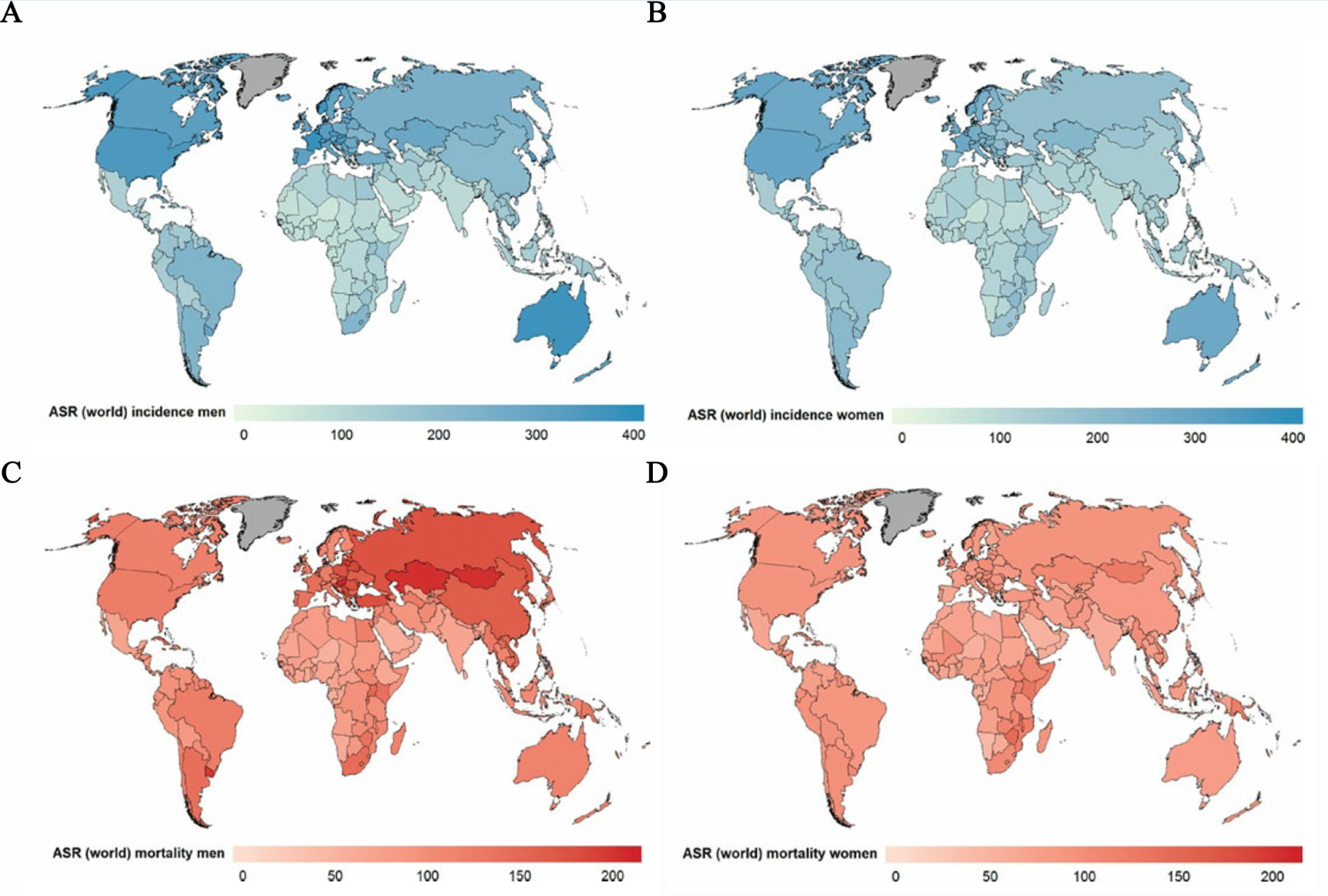
| + | cancers, due to their high incidence and serious mortality, are affecting | ||
| + | populations in all countries and all regions (Figure 1). However, in most | ||
| + | countries, resources for prevention and diagnosis of cancer still remain | ||
| + | limited due to their high cost and low cost-effectiveness</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">1</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">, whereas the early | ||
| + | detection of cancer has been proven to result in improved survival, less | ||
| + | extensive treatment and less possibility to metastasis</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">2-4</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">. Such situation | ||
| + | highlighted the undiminished importance of the development of a low-cost, | ||
| + | easily accessible and rapid tool for early screening and detection of cancers.</span> | ||
</p> | </p> | ||
| − | <p | + | <p style="text-indent:22pt;"> |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
</html> | </html> | ||
| − | [[File:T--NUDT CHINA--introfig1.jpg|700px|center ]] | + | [[File:T--NUDT CHINA--introfig1.jpg|700px|center]] |
<html> | <html> | ||
| − | |||
<p> | <p> | ||
| − | <b><span style="font-size: | + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Figure 1. |
| − | 1. Global distribution of estimated age-standardized world cancer incidence | + | Global distribution of estimated age-standardized world cancer incidence rate |
| − | rate (ASR) per 100 000 in (A) men, (B) women, and mortality rate (ASR) per 100 | + | (ASR) per 100 000 in (A) men, (B) women, and mortality rate (ASR) per 100 000 |
| − | 000 in (C) men and (D) women. (WHO, World cancer report, 2014)</span></b> | + | in (C) men and (D) women. (WHO, World cancer report, 2014)</span></b> |
</p> | </p> | ||
| − | <p align="center" style="text-align:center;"> | + | <p align="center" style="text-align:center;text-indent:22.1pt;"> |
| − | <b><span style="font-size:18px;"> </span></b> | + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span></b> |
</p> | </p> | ||
| − | <p> | + | <p style="text-indent:22pt;"> |
| − | <span style="font-size:18px;">MicroRNAs (miRNAs), as a kind of small | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">MicroRNAs |
| − | non-coding RNA containing approximately 22 nucleic acids, have been proven to | + | (miRNAs), as a kind of small non-coding RNA containing approximately 22 nucleic |
| − | play important roles on post-transcriptional regulation of the gene expression, | + | acids, have been proven to play important roles on post-transcriptional regulation |
| − | thus involving in the regulation of many important biological events</span><sup><span style="font-size:18px;">5</span></sup><span style="font-size:18px;">. | + | of the gene expression, thus involving in the regulation of many important |
| − | Recently, it was reported that serum miRNAs can serve as a promising cancer | + | biological events</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">5</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">. Recently, it was reported |
| − | biomarker because their expression pattern can be correlated with cancer type, | + | that serum miRNAs can serve as a promising cancer biomarker because their |
| − | stage, and other clinical variables, which then, implying that miRNA profiling | + | expression pattern can be correlated with cancer type, stage, and other |
| − | can be used as a tool for cancer diagnosis and prognosis</span><sup><span style="font-size:18px;">6-8</span></sup><span style="font-size:18px;">. | + | clinical variables, which then, implying that miRNA profiling can be used as a |
| − | Moreover, circulating miRNAs have been proven to remain stable under some | + | tool for cancer diagnosis and prognosis</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">6-8</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">. Moreover, circulating miRNAs |
| − | extreme condition such as RNase exposure, multiple freeze-thaw cycles, and | + | have been proven to remain stable under some extreme condition such as RNase |
| − | extreme pH, thus making them strong candidates for low-cost detection and | + | exposure, multiple freeze-thaw cycles, and extreme pH, thus making them strong |
| − | analysis </span><sup><span style="font-size:18px;">9</span></sup><span style="font-size:18px;">. | + | candidates for low-cost detection and analysis </span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">9</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">. However, due to their |
| − | However, due to their short length, low expression level and high homologous | + | short length, low expression level and high homologous sequence similarity, the |
| − | sequence similarity, the quantified detection and analyzation of circulating | + | quantified detection and analyzation of circulating miRNAs remain challenging |
| − | miRNAs remain challenging nowadays. Old-schools such as Northern Blotting, | + | nowadays. Old-schools such as Northern Blotting, microarray and qRT-PCR |
| − | microarray and qRT-PCR technique are still our approach to detect and analyze | + | technique are still our approach to detect and analyze the quantity of miRNA</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">10</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">. Notably, the expanded |
| − | the quantity of miRNA</span><sup><span style="font-size:18px;">10</span></sup><span style="font-size:18px;">. | + | application of these techniques, as well as some other new approaches such as |
| − | Notably, the expanded application of these techniques, as well as some other | + | bioluminescence</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">11</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">, Nanopore sensors</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">12</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;"> were severely limited |
| − | new approaches such as bioluminescence</span><sup><span style="font-size:18px;">11</span></sup><span style="font-size:18px;">, | + | due to their relatively low sensitivity (which were mostly nM sensitivity |
| − | Nanopore sensors</span><sup><span style="font-size:18px;">12</span></sup><span style="font-size:18px;"> were | + | |
| − | severely limited due to their relatively low sensitivity (which were mostly nM sensitivity | + | |
against the pM or even fM concentration of blood miRNA), cumbersome and complex | against the pM or even fM concentration of blood miRNA), cumbersome and complex | ||
| − | in operation, and relative high cost. More recently, Deng </span><i><span style="font-size:18px;">et.al </span></i><span style="font-size:18px;">reported a single-molecule resolution </span><i><span style="font-size:18px;">in situ</span></i><span style="font-size:18px;"> miRNA detection technique based on rolling circle | + | in operation, and relative high cost. More recently, Deng </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">et.al </span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;">reported a single-molecule resolution </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">in situ</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> miRNA detection technique based on rolling circle |
| − | amplification (RCA)</span><sup><span style="font-size:18px;">13</span></sup><span style="font-size:18px;">. | + | amplification (RCA)</span><sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">13</span></sup><span style="line-height:2;font-family:Perpetua;font-size:18px;">. However, this approach |
| − | However, this approach has been restricted only in the application of cellular </span><i><span style="font-size:18px;">in situ</span></i><span style="font-size:18px;"> analysis. Its expandability to | + | has been restricted only in the application of cellular </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">in situ</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> analysis. Its expandability to circulating miRNA detection |
| − | circulating miRNA detection still faces a major problem that the degree of one-step | + | still faces a major problem that the degree of one-step signal amplification and |
| − | signal amplification and differentiation might not be sufficient to meet the | + | differentiation might not be sufficient to meet the requirements of sensitivity |
| − | requirements of sensitivity and specificity. At the main time, such method | + | and specificity. At the main time, such method still relies on equipment such |
| − | still relies on equipment such as Fluoresce microplate readers or fluorescence | + | as Fluoresce microplate readers or fluorescence microscopes, which are highly |
| − | microscopes, which are highly costly.</span> | + | costly. </span> |
</p> | </p> | ||
| − | + | </br> | |
| − | + | ||
| − | </ | + | |
| − | + | ||
</html> | </html> | ||
| − | [[File:T--NUDT CHINA--introfig2.jpg|700px|center ]] | + | [[File:T--NUDT CHINA--introfig2.jpg|700px|center]] |
<html> | <html> | ||
| − | + | </br> | |
<p> | <p> | ||
| − | <b><span style="font-size: | + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Figure 2. |
| − | 2. Workflow for our CRISPR-based blood-microRNA detection system.</span></b> | + | Workflow for our CRISPR-based blood-microRNA detection system.</span></b> |
</p> | </p> | ||
<p> | <p> | ||
| − | <span style="font-size: | + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Using sequence information from online databases, |
| − | databases, probes for RCA reaction were designed in silico. Once synthesized | + | probes for RCA reaction were designed in silico. Once synthesized and sealed to |
| − | and sealed to form the dumbbell structure, the probe, together with other | + | form the dumbbell structure, the probe, together with other necessary materials |
| − | necessary materials can be embedded into tubes and freeze-dried to remain | + | can be embedded into tubes and freeze-dried to remain stable in room |
| − | stable in room temperature for a relatively long time. For the detection | + | temperature for a relatively long time. For the detection process, serum |
| − | process, serum samples were pre-treated by boiling in | + | samples were pre-treated by boiling in 95</span><span style="line-height:2;font-family:Perpetua;font-size:18px;">℃</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> for 15 min to expose |
| − | miRNAs completely. The amount of the specific RNA was indicated by a color | + | the miRNAs completely. The amount of the specific RNA was indicated by a color |
| − | difference in the tube from colorless to blue</span> | + | difference in the tube from colorless to blue</span> |
</p> | </p> | ||
| − | <p> | + | </br> |
| − | + | <p style="text-indent:22pt;"> | |
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In | ||
| + | our project, we designed a novel cell-free platform built with synthetic | ||
| + | bio-components to achieve the low-cost, handy and visualized detection of serum | ||
| + | miRNAs, which can be employed in low-resource settings (Figure 2). Using miR | ||
| + | let-7a (a bio-marker for non-small cell lung cancer (NSCLC)) as a demo of our | ||
| + | scheme, we modified the RCA based DNA amplification system and introduced it | ||
| + | into nucleic acid detection in liquid samples such as serum, and then conducted | ||
| + | Sybr I mediated fluorescent assay for its validation and assessment. The | ||
| + | improvements of sensitivity and specificity of RCA output signal as well as the | ||
| + | visualization of RCA outputs were achieved through a single guide RNA (sgRNA) | ||
| + | mediated dCas9 binding system and a conjugated split-HRP reporting system. | ||
| + | Meanwhile, a mathematic model was also developed to provide theoretical | ||
| + | approval to our scheme and basic guideline for wet-lab experiments. Finally, we | ||
| + | employed a simple sample-pretreatment protocol to reliably expose miRNAs in | ||
| + | serum samples and demonstrated robust detection with this scheme to compare | ||
| + | let-7a concentrations among blood samples collected from NSCLC patients and healthy | ||
| + | volunteers. </span> | ||
</p> | </p> | ||
| − | <p | + | <p style="text-indent:22pt;"> |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <span style="font-size:18px;"> </span> | + | |
</p> | </p> | ||
<h2> | <h2> | ||
| − | + | <span><span style="color:#7f1015">Improvement We Made</span></span><hr /> | |
| − | + | ||
</h2> | </h2> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"><strong>BBa_K1789003 and BBa_K1789004</strong></span> | |
</p> | </p> | ||
| − | <p> | + | <p style="text-indent:22pt;"> |
| − | <span style="font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In |
| + | our project this year, a new protein-protein interaction (PPI) toolkit | ||
| + | containing several split reporting systems were modified and designed and | ||
| + | introduced into the registry. As a classical PPI indicator, split-GFP system, | ||
| + | developed previously in our project in iGEM2015 (BBa_K1789003 and | ||
| + | BBa_K1789004), was also included in our kit. Several improvements has been made | ||
| + | for this system including:</span> | ||
</p> | </p> | ||
| − | <p> | + | <p style="text-indent:-0.55pt;"> |
| − | <span style="font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1.</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Improved |
| − | + | characterization for previous parts</span> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
| − | <p style="text-indent: | + | <p style="text-indent:22pt;"> |
| − | <span style="font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To |
| − | + | further improve the function of existing parts, we stimulate an </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">in vivo</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> PPI situation, and tried to | |
| + | optimize the culture condition for a better signal-to-noise ratio (SNR). For | ||
| + | such matter, two devices, containing split-GFP fragments and a complete or | ||
| + | spited zinc finger protein, were built under control of a lac operon controlled | ||
| + | T7 promoter. The complete zinc finger protein was to stimulate a PPI positive | ||
| + | situation, while the split one was to stimulate a PPI negative situation | ||
| + | (Figure 3A). Fluorescence signal was detected by a microplate reader after an | ||
| + | overnight culture under various conditions.</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Relative fluorescence intensity was then calculated with normalization | ||
| + | of OD</span><sub><span style="line-height:2;font-family:Perpetua;font-size:18px;">600</span></sub><span style="line-height:2;font-family:Perpetua;font-size:18px;"> value. The relative fluorescence intensity of each control | ||
| + | group was set arbitrarily at 1.0 (data not shown), and the levels of the other | ||
| + | groups were adjusted correspondingly. Results shown a better SNR under 20</span><span style="line-height:2;font-family:Perpetua;font-size:18px;">℃</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> and 0.5mM IPTG induction (Figure 3B). Thus indicating that better performance | ||
| + | of such system could be expected under lower culturing temperature.</span> | ||
</p> | </p> | ||
| − | <p | + | <p style="text-indent:22pt;"> |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
<p> | <p> | ||
| − | <span style="font-size:18px;"> | + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Figure |
| − | + | 3 The green fluorescence (Ex: 488 nm; Em: 538 nm) of split GFP after overnight | |
| − | + | culture of E. coli with or without IPTG induction under different express | |
| − | + | condition. </span></b> | |
</p> | </p> | ||
<p> | <p> | ||
| − | <span style="font-size:18px;">& | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Relative fluorescence intensity was |
| + | calculated with normalization of the OD600 value. The relative fluorescence | ||
| + | intensity of each control group was set arbitrarily at 1.0 (data not shown), | ||
| + | and the levels of the other groups were adjusted correspondingly. This | ||
| + | experiment was run in three parallel reactions, and the data represent results | ||
| + | obtained from at least three independent experiments. *p<0.05, **p<0.01.</span> | ||
</p> | </p> | ||
| − | <p | + | <p style="text-indent:22pt;"> |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | |
</p> | </p> | ||
| − | <p | + | <p style="text-indent:22pt;"> |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | |
| − | + | ||
| − | + | ||
| − | <span style="font-size:18px;"> </span> | + | |
</p> | </p> | ||
| − | <p style="text-indent: | + | <p style="text-indent:6.5pt;"> |
| − | <span style="font-size:18px;">2. To | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">2.</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">To |
further improve the function of split-GFP system, another method of splitting | further improve the function of split-GFP system, another method of splitting | ||
GFP was introduced and tested in our project. Instead of a traditional two-part | GFP was introduced and tested in our project. Instead of a traditional two-part | ||
split, we split the GFP protein into three fragments namely GFP10 (residues | split, we split the GFP protein into three fragments namely GFP10 (residues | ||
| − | 194-212), GFP11 (residues 213-233) and GFP 1-9 (residues 1-193) [sci-reps 10.103/srep02854]. Due to their short | + | 194-212), GFP11 (residues 213-233) and GFP 1-9 (residues 1-193) </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">[sci-reps 10.103/srep02854]</span><span style="line-height:2;font-family:Perpetua;font-size:18px;">. Due to their short |
length, two small fragments can be easily fused onto proteins with less | length, two small fragments can be easily fused onto proteins with less | ||
| − | affection on their folding (figure 4A).</span> | + | affection on their folding </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">(figure 4A)</span><span style="line-height:2;font-family:Perpetua;font-size:18px;">. </span> |
</p> | </p> | ||
<p> | <p> | ||
| − | <span style="font-size:18px;"> </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> |
</p> | </p> | ||
| − | <p align="center" style="text-align:center;"> | + | <p align="center" style="text-align:center;text-indent:22.1pt;"> |
| − | <b><span style="font-size: | + | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">(Figure |
| + | 4)</span></b> | ||
</p> | </p> | ||
| − | <p | + | <p style="text-indent:22pt;"> |
| − | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> | |
</p> | </p> | ||
| − | <p> | + | <p style="text-indent:22pt;"> |
| − | <span style="font-size:18px;">Comparing with previous | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Comparing |
| − | split-GFP system, higher SNR was reached under the same expression condition, | + | with previous split-GFP system, higher SNR was reached under the same |
| − | while the total signal intensity suffered tolerable decrease (Figure 4B).</span> | + | expression condition, while the total signal intensity suffered tolerable |
| + | decrease (Figure 4B).</span> | ||
</p> | </p> | ||
<p> | <p> | ||
| − | <span style="font-size:18px;"> </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> |
</p> | </p> | ||
<h2> | <h2> | ||
| − | + | <span><span style="color:#7f1015">Reference</span></span><hr /> | |
</h2> | </h2> | ||
| − | |||
| − | |||
| − | |||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">1 Jeong, | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Jeong, |
K. E. & Cairns, J. A. Review of economic evidence in the prevention and | K. E. & Cairns, J. A. Review of economic evidence in the prevention and | ||
| − | early detection of colorectal cancer. </span><i><span style="font-size:18px;">Health | + | early detection of colorectal cancer. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Health |
| − | Econ Rev</span></i> <b><span style="font-size:18px;">3</span></b><span style="font-size:18px;">, 20, | + | Econ Rev</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">3</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 20, |
| − | doi:10.1186/2191-1991-3-20 (2013).</span> | + | doi:10.1186/2191-1991-3-20 (2013).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">2 Etzioni, R.</span><i><span style="font-size:18px;"> et al.</span></i><span style="font-size:18px;"> The case for early detection. </span><i><span style="font-size:18px;">Nat Rev Cancer</span></i> <b><span style="font-size:18px;">3</span></b><span style="font-size:18px;">, | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">2</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Etzioni, R.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> The case for early detection. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Nat Rev Cancer</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">3</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, |
| − | 243-252, doi:10.1038/nrc1041 (2003).</span> | + | 243-252, doi:10.1038/nrc1041 (2003).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">3 Wolf, A. M.</span><i><span style="font-size:18px;"> et al.</span></i><span style="font-size:18px;"> American Cancer Society guideline for the early detection | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">3</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Wolf, A. M.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> American Cancer Society guideline for the early detection |
| − | of prostate cancer: update 2010. </span><i><span style="font-size:18px;">CA | + | of prostate cancer: update 2010. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">CA |
| − | Cancer J Clin</span></i> <b><span style="font-size:18px;">60</span></b><span style="font-size:18px;">, 70-98, | + | Cancer J Clin</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">60</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 70-98, |
| − | doi:10.3322/caac.20066 (2010).</span> | + | doi:10.3322/caac.20066 (2010).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">4 McPhail, S., Johnson, S., Greenberg, D., | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">4</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">McPhail, S., Johnson, S., Greenberg, |
| − | Peake, M. & Rous, B. Stage at diagnosis and early mortality from cancer | + | D., Peake, M. & Rous, B. Stage at diagnosis and early mortality from cancer |
| − | England. </span><i><span style="font-size:18px;">Br J Cancer</span></i> <b><span style="font-size:18px;">112 Suppl 1</span></b><span style="font-size:18px;">, S108-115, | + | in England. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Br J Cancer</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">112 Suppl 1</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, S108-115, |
| − | doi:10.1038/bjc.2015.49 (2015).</span> | + | doi:10.1038/bjc.2015.49 (2015).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">5 Ambros, V. MicroRNA pathways in flies | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">5</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Ambros, V. MicroRNA pathways in flies |
| − | worms: growth, death, fat, stress, and timing. </span><i><span style="font-size:18px;">Cell</span></i> <b><span style="font-size:18px;">113</span></b><span style="font-size:18px;">, 673-676 | + | and worms: growth, death, fat, stress, and timing. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Cell</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">113</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 673-676 |
| − | (2003).</span> | + | (2003).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">6 Lu, J.</span><i><span style="font-size:18px;"> et al.</span></i><span style="font-size:18px;"> MicroRNA expression profiles classify human cancers. </span><i><span style="font-size:18px;">Nature</span></i> <b><span style="font-size:18px;">435</span></b><span style="font-size:18px;">, 834-838, doi:10.1038/nature03702 (2005).</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">6</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Lu, J.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> MicroRNA expression profiles classify human cancers. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Nature</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">435</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 834-838, doi:10.1038/nature03702 (2005).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">7 Jansson, M. D. & Lund, A. H. | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">7</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Jansson, M. D. & Lund, A. H. |
| − | and cancer. </span><i><span style="font-size:18px;">Mol Oncol</span></i> <b><span style="font-size:18px;">6</span></b><span style="font-size:18px;">, 590-610, | + | MicroRNA and cancer. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Mol Oncol</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">6</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 590-610, |
| − | doi:10.1016/j.molonc.2012.09.006 (2012).</span> | + | doi:10.1016/j.molonc.2012.09.006 (2012).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">8 Schultz, N. A.</span><i><span style="font-size:18px;"> et al.</span></i><span style="font-size:18px;"> MicroRNA biomarkers in whole blood for detection of | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">8</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Schultz, N. A.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> MicroRNA biomarkers in whole blood for detection of |
| − | pancreatic cancer. </span><i><span style="font-size:18px;">JAMA</span></i> <b><span style="font-size:18px;">311</span></b><span style="font-size:18px;">, 392-404, | + | pancreatic cancer. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">JAMA</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">311</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 392-404, |
| − | doi:10.1001/jama.2013.284664 (2014).</span> | + | doi:10.1001/jama.2013.284664 (2014).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">9 Chen, X.</span><i><span style="font-size:18px;"> et al.</span></i><span style="font-size:18px;"> Characterization of microRNAs in serum: a novel class of | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">9</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Chen, X.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> Characterization of microRNAs in serum: a novel class of |
| − | biomarkers for diagnosis of cancer and other diseases. </span><i><span style="font-size:18px;">Cell Res</span></i> <b><span style="font-size:18px;">18</span></b><span style="font-size:18px;">, 997-1006, | + | biomarkers for diagnosis of cancer and other diseases. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Cell Res</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">18</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 997-1006, |
| − | doi:10.1038/cr.2008.282 (2008).</span> | + | doi:10.1038/cr.2008.282 (2008).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">10 Hunt, E. A., Broyles, D., Head, T. & | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">10</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Hunt, E. A., Broyles, D., Head, T. & |
| − | Deo, S. K. MicroRNA Detection: Current Technology and Research Strategies. </span><i><span style="font-size:18px;">Annu Rev Anal Chem (Palo Alto Calif)</span></i> <b><span style="font-size:18px;">8</span></b><span style="font-size:18px;">, 217-237, doi:10.1146/annurev-anchem-071114-040343 | + | Deo, S. K. MicroRNA Detection: Current Technology and Research Strategies. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Annu Rev Anal Chem (Palo Alto Calif)</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">8</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 217-237, |
| − | (2015).</span> | + | doi:10.1146/annurev-anchem-071114-040343 (2015).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">11 Cissell, K. A., Rahimi, Y., Shrestha, S., | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">11</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Cissell, K. A., Rahimi, Y., Shrestha, |
| − | Hunt, E. A. & Deo, S. K. Bioluminescence-based detection of microRNA, | + | S., Hunt, E. A. & Deo, S. K. Bioluminescence-based detection of microRNA, |
| − | in breast cancer cells. </span><i><span style="font-size:18px;">Anal Chem</span></i> <b><span style="font-size:18px;">80</span></b><span style="font-size:18px;">, 2319-2325, doi:10.1021/ac702577a | + | miR21 in breast cancer cells. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Anal Chem</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">80</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 2319-2325, doi:10.1021/ac702577a |
| − | (2008).</span> | + | (2008).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">12 Venkatesan, B. M. & Bashir, R. | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">12</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Venkatesan, B. M. & Bashir, R. |
| − | sensors for nucleic acid analysis. </span><i><span style="font-size:18px;">Nature | + | Nanopore sensors for nucleic acid analysis. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Nature |
| − | nanotechnology</span></i> <b><span style="font-size:18px;">6</span></b><span style="font-size:18px;">, 615-624, | + | nanotechnology</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">6</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, 615-624, |
| − | doi:10.1038/nnano.2011.129 (2011).</span> | + | doi:10.1038/nnano.2011.129 (2011).</span> |
</p> | </p> | ||
<p style="text-indent:-36pt;"> | <p style="text-indent:-36pt;"> | ||
| − | <span style="font-size:18px;">13 Deng, R.</span><i><span style="font-size:18px;"> et al.</span></i><span style="font-size:18px;"> Toehold-initiated rolling circle amplification for | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">13</span><span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span><span style="line-height:2;font-family:Perpetua;font-size:18px;">Deng, R.</span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> et al.</span></i><span style="line-height:2;font-family:Perpetua;font-size:18px;"> Toehold-initiated rolling circle amplification for |
| − | individual microRNAs in situ in single cells. </span><i><span style="font-size:18px;">Angew Chem Int Ed Engl</span></i> <b><span style="font-size:18px;">53</span></b><span style="font-size:18px;">, | + | visualizing individual microRNAs in situ in single cells. </span><i><span style="line-height:2;font-family:Perpetua;font-size:18px;">Angew Chem Int Ed Engl</span></i><span> </span><b><span style="line-height:2;font-family:Perpetua;font-size:18px;">53</span></b><span style="line-height:2;font-family:Perpetua;font-size:18px;">, |
| − | 2389-2393, doi:10.1002/anie.201309388 (2014).</span> | + | 2389-2393, doi:10.1002/anie.201309388 (2014).</span> |
</p> | </p> | ||
<p> | <p> | ||
| − | <span style="font-size:18px;"> </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> </span> |
</p> | </p> | ||
| − | |||
Revision as of 02:21, 14 October 2016